Applications of Lgr5-Positive Cochlear Progenitors (LCPs) to the Study of Hair Cell Differentiation
- PMID: 30873406
- PMCID: PMC6401656
- DOI: 10.3389/fcell.2019.00014
Applications of Lgr5-Positive Cochlear Progenitors (LCPs) to the Study of Hair Cell Differentiation
Abstract
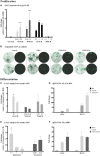
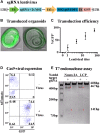
The mouse cochlea contains approximately 15,000 hair cells. Its dimensions and location, and the small number of hair cells, make mechanistic, developmental and cellular replacement studies difficult. We recently published a protocol to expand and differentiate murine neonatal cochlear progenitor cells into 3D organoids that recapitulate developmental pathways and can generate large numbers of hair cells with intact stereociliary bundles, molecular markers of the native cells and mechanotransduction channel activity, as indicated by FM1-43 uptake. Here, we elaborate on the method and application of these Lgr5-positive cochlear progenitors, termed LCPs, to the study of inner ear development and differentiation. We demonstrate the use of these cells for testing several drug candidates, gene silencing and overexpression, as well as genomic modification using CRISPR/Cas9. We thus establish LCPs as a valuable in vitro tool for the analysis of progenitor cell manipulation and hair cell differentiation.
Keywords: Lgr5; cochlea; differentiation; epigenetics; hair cells; proliferation; supporting cells.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

