Omics and the Search for Blood Biomarkers in Chronic Obstructive Pulmonary Disease. Insights from COPDGene
- PMID: 30874442
- PMCID: PMC6670029
- DOI: 10.1165/rcmb.2018-0245PS
Omics and the Search for Blood Biomarkers in Chronic Obstructive Pulmonary Disease. Insights from COPDGene
Abstract
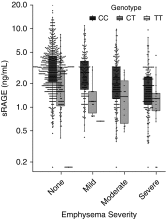
There is an unmet need for blood biomarkers in diagnosis and prognosis of chronic obstructive pulmonary disease (COPD). The search for these biomarkers has been revolutionized by high-throughput sequencing techniques and multiplex platforms that can measure thousands of gene transcripts, proteins, or metabolites. We review COPDGene (Genetic Epidemiology of COPD) project publications that include DNA methylation, transcriptomic, proteomic, and metabolomic blood biomarkers and discuss their impact on COPD. Key contributions from COPDGene include identification of DNA methylation effects from smoking and genetic variation, new transcriptomic signatures in the blood, identification of protein biomarkers associated with severity and progression (e.g., sRAGE [soluble receptor for advanced glycosylation end products], inflammatory cytokines IL-6 and IL-8), and identification of small molecules (ceramides and sphingomyelin) that may be pathogenic. COPDGene studies have revealed that some of the COPD genome-wide association study polymorphisms are strongly associated with blood biomarkers (e.g., rs2070600 in AGER is a pQTL [protein quantitative trait locus] for sRAGE), underscoring the importance of combining omics results. Investigators have developed molecular networks identifying lower CD4+ resting memory cells associated with COPD. Genes, proteins, and metabolite networks are particularly important because the explanatory value of any single molecule is small (1-10%) compared with panels of multiple markers. COPDGene has been a useful resource in the identification and validation of multiple biomarkers for COPD. These biomarkers, either combined in multiple biomarker panels or integrated with other omics data types, may lead to novel diagnostic and prognostic tests for COPD phenotypes and may be relevant for assessing novel therapies.
Keywords: biomarkers; chronic obstructive pulmonary disease; epigenetics; gene expression; metabolomics.
Figures

References
-
- Agusti A, Soriano JB. COPD as a systemic disease. COPD. 2008;5:133–138. - PubMed
-
- Müllerova H, Agusti A, Erqou S, Mapel DW. Cardiovascular comorbidity in COPD: systematic literature review. Chest. 2013;144:1163–1178. - PubMed
-
- Faner R, Tal-Singer R, Riley JH, Celli B, Vestbo J, MacNee W, et al. ECLIPSE Study Investigators. Lessons from ECLIPSE: a review of COPD biomarkers. Thorax. 2014;69:666–672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

