Automating multimodal microscopy with NanoJ-Fluidics
- PMID: 30874553
- PMCID: PMC6420627
- DOI: 10.1038/s41467-019-09231-9
Automating multimodal microscopy with NanoJ-Fluidics
Abstract
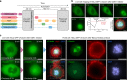
Combining and multiplexing microscopy approaches is crucial to understand cellular events, but requires elaborate workflows. Here, we present a robust, open-source approach for treating, labelling and imaging live or fixed cells in automated sequences. NanoJ-Fluidics is based on low-cost Lego hardware controlled by ImageJ-based software, making high-content, multimodal imaging easy to implement on any microscope with high reproducibility. We demonstrate its capacity on event-driven, super-resolved live-to-fixed and multiplexed STORM/DNA-PAINT experiments.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

