A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum
- PMID: 30874562
- PMCID: PMC6420630
- DOI: 10.1038/s41467-019-09145-6
A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum
Abstract
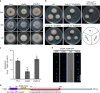
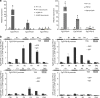
Sterol biosynthesis is controlled by transcription factor SREBP in many eukaryotes. Here, we show that SREBP orthologs are not involved in the regulation of sterol biosynthesis in Fusarium graminearum, a fungal pathogen of cereal crops worldwide. Instead, sterol production is controlled in this organism by a different transcription factor, FgSR, that forms a homodimer and binds to a 16-bp cis-element of its target gene promoters containing two conserved CGAA repeat sequences. FgSR is phosphorylated by the MAP kinase FgHog1, and the phosphorylated FgSR interacts with the chromatin remodeling complex SWI/SNF at the target genes, leading to enhanced transcription. Interestingly, FgSR orthologs exist only in Sordariomycetes and Leotiomycetes fungi. Additionally, FgSR controls virulence mainly via modulating deoxynivalenol biosynthesis and responses to phytoalexin.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

