CyTOFmerge: integrating mass cytometry data across multiple panels
- PMID: 30874801
- PMCID: PMC6792069
- DOI: 10.1093/bioinformatics/btz180
CyTOFmerge: integrating mass cytometry data across multiple panels
Abstract
Motivation: High-dimensional mass cytometry (CyTOF) allows the simultaneous measurement of multiple cellular markers at single-cell level, providing a comprehensive view of cell compositions. However, the power of CyTOF to explore the full heterogeneity of a biological sample at the single-cell level is currently limited by the number of markers measured simultaneously on a single panel.
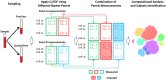
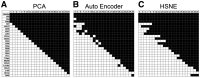
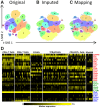
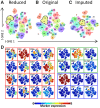
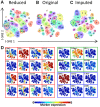
Results: To extend the number of markers per cell, we propose an in silico method to integrate CyTOF datasets measured using multiple panels that share a set of markers. Additionally, we present an approach to select the most informative markers from an existing CyTOF dataset to be used as a shared marker set between panels. We demonstrate the feasibility of our methods by evaluating the quality of clustering and neighborhood preservation of the integrated dataset, on two public CyTOF datasets. We illustrate that by computationally extending the number of markers we can further untangle the heterogeneity of mass cytometry data, including rare cell-population detection.
Availability and implementation: Implementation is available on GitHub (https://github.com/tabdelaal/CyTOFmerge).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Bandura D.R. et al. (2009) Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem., 81, 6813–6822. - PubMed

