Systematic analysis of Type I-E Escherichia coli CRISPR-Cas PAM sequences ability to promote interference and primed adaptation
- PMID: 30875129
- PMCID: PMC6568314
- DOI: 10.1111/mmi.14237
Systematic analysis of Type I-E Escherichia coli CRISPR-Cas PAM sequences ability to promote interference and primed adaptation
Abstract
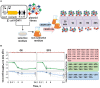
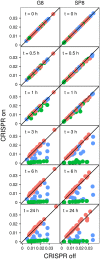
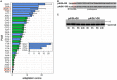
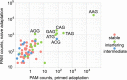
CRISPR interference occurs when a protospacer recognized by the CRISPR RNA is destroyed by Cas effectors. In Type I CRISPR-Cas systems, protospacer recognition can lead to «primed adaptation» - acquisition of new spacers from in cis located sequences. Type I CRISPR-Cas systems require the presence of a trinucleotide protospacer adjacent motif (PAM) for efficient interference. Here, we investigated the ability of each of 64 possible trinucleotides located at the PAM position to induce CRISPR interference and primed adaptation by the Escherichia coli Type I-E CRISPR-Cas system. We observed clear separation of PAM variants into three groups: those unable to cause interference, those that support rapid interference and those that lead to reduced interference that occurs over extended periods of time. PAM variants unable to support interference also did not support primed adaptation; those that supported rapid interference led to no or low levels of adaptation, while those that caused attenuated levels of interference consistently led to highest levels of adaptation. The results suggest that primed adaptation is fueled by the products of CRISPR interference. Extended over time interference with targets containing «attenuated» PAM variants provides a continuous source of new spacers leading to high overall level of spacer acquisition.
© 2019 The Authors. Molecular Microbiology Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declares that there is no conflict of interest regarding the publication of this article.
Figures

Similar articles
-
Avoidance of Trinucleotide Corresponding to Consensus Protospacer Adjacent Motif Controls the Efficiency of Prespacer Selection during Primed Adaptation.mBio. 2018 Dec 4;9(6):e02169-18. doi: 10.1128/mBio.02169-18. mBio. 2018. PMID: 30514784 Free PMC article.
-
Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7626-31. doi: 10.1073/pnas.1602639113. Epub 2016 Jun 20. Proc Natl Acad Sci U S A. 2016. PMID: 27325762 Free PMC article.
-
High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli.RNA Biol. 2013 May;10(5):716-25. doi: 10.4161/rna.24325. Epub 2013 Apr 25. RNA Biol. 2013. PMID: 23619643 Free PMC article.
-
CRISPR-Cas adaptive immune systems in Sulfolobales: genetic studies and molecular mechanisms.Sci China Life Sci. 2021 May;64(5):678-696. doi: 10.1007/s11427-020-1745-0. Epub 2020 Oct 29. Sci China Life Sci. 2021. PMID: 33140167 Review.
-
Diversity of CRISPR-Cas-Mediated Mechanisms of Adaptive Immunity in Prokaryotes and Their Application in Biotechnology.Biochemistry (Mosc). 2016 Jul;81(7):653-61. doi: 10.1134/S0006297916070026. Biochemistry (Mosc). 2016. PMID: 27449612 Review.
Cited by
-
Dependence of post-segregational killing mediated by Type II restriction-modification systems on the lifetime of restriction endonuclease effective activity.mBio. 2024 Aug 14;15(8):e0140824. doi: 10.1128/mbio.01408-24. Epub 2024 Jul 9. mBio. 2024. PMID: 38980007 Free PMC article.
-
CRISPR-Cas adaptation in Escherichia coli.Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review.
-
Identification of Spacer and Protospacer Sequence Requirements in the Vibrio cholerae Type I-E CRISPR/Cas System.mSphere. 2020 Nov 18;5(6):e00813-20. doi: 10.1128/mSphere.00813-20. mSphere. 2020. PMID: 33208517 Free PMC article.
-
Single cell variability of CRISPR-Cas interference and adaptation.Mol Syst Biol. 2022 Apr;18(4):e10680. doi: 10.15252/msb.202110680. Mol Syst Biol. 2022. PMID: 35467080 Free PMC article.
-
PAM-repeat associations and spacer selection preferences in single and co-occurring CRISPR-Cas systems.Genome Biol. 2021 Sep 30;22(1):281. doi: 10.1186/s13059-021-02495-9. Genome Biol. 2021. PMID: 34593010 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

