Environmentally-induced epigenetic conversion of a piRNA cluster
- PMID: 30875295
- PMCID: PMC6420265
- DOI: 10.7554/eLife.39842
Environmentally-induced epigenetic conversion of a piRNA cluster
Abstract
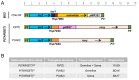
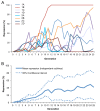
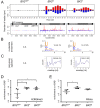
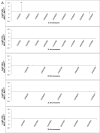
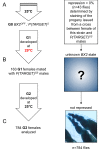
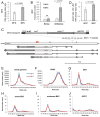
Transposable element (TE) activity is repressed in animal gonads by PIWI-interacting RNAs (piRNAs) produced by piRNA clusters. Current models in flies propose that germinal piRNA clusters are functionally defined by the maternal inheritance of piRNAs produced during the previous generation. Taking advantage of an inactive, but ready to go, cluster of P-element derived transgene insertions in Drosophila melanogaster, we show here that raising flies at high temperature (29°C) instead of 25°C triggers the stable conversion of this locus from inactive into actively producing functional piRNAs. The increase of antisense transcripts from the cluster at 29°C combined with the requirement of transcription of euchromatic homologous sequences, suggests a role of double stranded RNA in the production of de novo piRNAs. This report describes the first case of the establishment of an active piRNA cluster by environmental changes in the absence of maternal inheritance of homologous piRNAs.
Editorial note: This article has been through an editorial process in which the authors decide how to respond to the issues raised during peer review. The Reviewing Editor's assessment is that all the issues have been addressed (see decision letter).
Keywords: D. melanogaster; epigenetics; genetics; genomics; germline; heterochromatin; non coding small RNA; temperature; transposable element.
© 2019, Casier et al.
Conflict of interest statement
KC, VD, NG, CH, EV, CV, SR, EB, LT, AB No competing interests declared
Figures








References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

