Function and regulation of chromatin insulators in dynamic genome organization
- PMID: 30875678
- PMCID: PMC6692201
- DOI: 10.1016/j.ceb.2019.02.001
Function and regulation of chromatin insulators in dynamic genome organization
Abstract
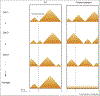
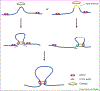
Chromatin insulators are DNA-protein complexes that play a crucial role in regulating chromatin organization. Within the past two years, a plethora of genome-wide conformation capture studies have helped reveal that insulators are necessary for proper genome-wide organization of topologically associating domains, which are formed in a manner distinct from that of compartments. These studies have also provided novel insights into the mechanics of how CTCF/cohesin-dependent loops form in mammals, strongly supporting the loop extrusion model. In combination with single-cell imaging approaches in both Drosophila and mammals, the dynamics of insulator-mediated chromatin interactions are also coming to light. Insulator-dependent structures vary across individual cells and tissues, highlighting the need to study the regulation of insulators in particular temporal and spatial contexts throughout development.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of Interest
The authors declare no conflict of interest.
Figures
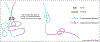

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

