Genomic analysis demonstrates that histologically-defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis
- PMID: 30876455
- PMCID: PMC6419470
- DOI: 10.1186/s40478-019-0689-3
Genomic analysis demonstrates that histologically-defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis
Abstract
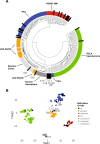
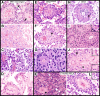
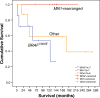
Astroblastoma (AB) is a rare CNS tumor demonstrating abundant astroblastomatous pseudorosettes. Its molecular features have not been comprehensively studied and its status as a tumor entity is controversial. We analyzed a cohort of 27 histologically-defined ABs using DNA methylation profiling, copy number analysis, FISH and site-directed sequencing. Most cases demonstrated mutually exclusive MN1 rearrangements (n = 10) or BRAFV600E mutations (n = 7). Two additional cases harbored RELA rearrangements. Other cases lacked these specific genetic alterations (n = 8). By DNA methylation profiling, tumors with MN1 or RELA rearrangement clustered with high-grade neuroepithelial tumor with MN1 alteration (HGNET-MN1) and RELA-fusion ependymoma, respectively. In contrast, BRAFV600E-mutant tumors grouped with pleomorphic xanthoastrocytoma (PXA). Six additional tumors clustered with either supratentorial pilocytic astrocytoma and ganglioglioma (LGG-PA/GG-ST), normal or reactive cerebrum, or with no defined DNA methylation class. While certain histologic features favored one genetic group over another, no group could be reliably distinguished by histopathology alone. Survival analysis between genetic AB subtypes was limited by sample size, but showed that MN1-rearranged AB tumors were characterized by better overall survival compared to other genetic subtypes, in fact, significantly better than BRAFV600E-mutant tumors (P = 0.013). Our data confirm that histologically-defined ABs are molecularly heterogeneous and do not represent a single entity. They rather encompass several low- to higher-grade glial tumors including neuroepithelial tumors with MN1 rearrangement, PXA-like tumors, RELA ependymomas, and possibly yet uncharacterized lesions. Genetic subtyping of tumors exhibiting AB histology, particularly determination of MN1 and BRAFV600E status, is necessary for important prognostic and possible treatment implications.
Conflict of interest statement
Ethics approval and consent to participate
Approval for the study of human tissue was obtained by the Institutional Review Board of each participating institution.
Consent for publication
No patient personal identifying data is included in the study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Aldape KD, Rosenblum MK. Astroblastoma. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. Lyon, France: International Agency for Research on Cancer; 2016. pp. 121–122.
-
- Broniscer A, Chamdine O, Hwang S, Lin T, Pounds S, Onar-Thomas A, Shurtleff S, Allen S, Gajjar A, Northcott P, et al. Gliomatosis cerebri in children shares molecular characteristics with other pediatric gliomas. Acta Neuropathol. 2016;131:299–307. doi: 10.1007/s00401-015-1532-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

