Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis
- PMID: 30876876
- PMCID: PMC6477154
- DOI: 10.1016/j.immuni.2019.02.006
Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis
Abstract
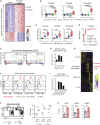
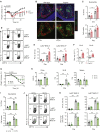
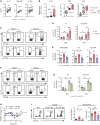
Inflammatory bowel disease is a chronic, relapsing condition with two subtypes, Crohn's disease (CD) and ulcerative colitis (UC). Genome-wide association studies (GWASs) in UC implicate a FCGR2A variant that alters the binding affinity of the antibody receptor it encodes, FcγRIIA, for immunoglobulin G (IgG). Here, we aimed to understand the mechanisms whereby changes in FcγRIIA affinity would affect inflammation in an IgA-dominated organ. We found a profound induction of anti-commensal IgG and a concomitant increase in activating FcγR signaling in the colonic mucosa of UC patients. Commensal-IgG immune complexes engaged gut-resident FcγR-expressing macrophages, inducing NLRP3- and reactive-oxygen-species-dependent production of interleukin-1β (IL-1β) and neutrophil-recruiting chemokines. These responses were modulated by the FCGR2A genotype. In vivo manipulation of macrophage FcγR signal strength in a mouse model of UC determined the magnitude of intestinal inflammation and IL-1β-dependent type 17 immunity. The identification of an important contribution of IgG-FcγR-dependent inflammation to UC has therapeutic implications.
Keywords: Fcγ receptors; IL-1β; IgG; inflammatory bowel disease; type 17 immunity.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Becalming Type 17 Inflammation in Ulcerative Colitis.Immunity. 2019 Apr 16;50(4):1029-1031. doi: 10.1016/j.immuni.2019.03.019. Immunity. 2019. PMID: 30995495
Dataset use reported in
-
Becalming Type 17 Inflammation in Ulcerative Colitis.Immunity. 2019 Apr 16;50(4):1029-1031. doi: 10.1016/j.immuni.2019.03.019. Immunity. 2019. PMID: 30995495
References
-
- Abraham C., Cho J.H. Functional consequences of NOD2 (CARD15) mutations. Inflamm. Bowel Dis. 2006;12:641–650. - PubMed
-
- Arijs I., Li K., Toedter G., Quintens R., Van Lommel L., Van Steen K., Leemans P., De Hertogh G., Lemaire K., Ferrante M. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58:1612–1619. - PubMed
-
- Asano K., Matsushita T., Umeno J., Hosono N., Takahashi A., Kawaguchi T., Matsumoto T., Matsui T., Kakuta Y., Kinouchi Y. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 2009;41:1325–1329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

