Imperfect Linkage Disequilibrium Generates Phantom Epistasis (& Perils of Big Data)
- PMID: 30877081
- PMCID: PMC6505142
- DOI: 10.1534/g3.119.400101
Imperfect Linkage Disequilibrium Generates Phantom Epistasis (& Perils of Big Data)
Abstract
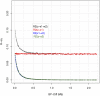
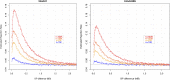
The genetic architecture of complex human traits and diseases is affected by large number of possibly interacting genes, but detecting epistatic interactions can be challenging. In the last decade, several studies have alluded to problems that linkage disequilibrium can create when testing for epistatic interactions between DNA markers. However, these problems have not been formalized nor have their consequences been quantified in a precise manner. Here we use a conceptually simple three locus model involving a causal locus and two markers to show that imperfect LD can generate the illusion of epistasis, even when the underlying genetic architecture is purely additive. We describe necessary conditions for such "phantom epistasis" to emerge and quantify its relevance using simulations. Our empirical results demonstrate that phantom epistasis can be a very serious problem in GWAS studies (with rejection rates against the additive model greater than 0.28 for nominal p-values of 0.05, even when the model is purely additive). Some studies have sought to avoid this problem by only testing interactions between SNPs with R-sq. <0.1. We show that this threshold is not appropriate and demonstrate that the magnitude of the problem is even greater with large sample size, intermediate allele frequencies, and when the causal locus explains a large amount of phenotypic variance. We conclude that caution must be exercised when interpreting GWAS results derived from very large data sets showing strong evidence in support of epistatic interactions between markers.
Keywords: Big Data; GWAS; apparent epistasis; epistasis; imperfect LD; linkage disequilibrium; missing heritability; phantom epistasis.
Copyright © 2019 de los Campos et al.
Figures


References
-
- Bulmer M. G., 1971. The Effect of Selection on Genetic Variability. Am. Nat. 105: 201–211. 10.1086/282718 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
