Cleaning Genotype Data from Diversity Outbred Mice
- PMID: 30877082
- PMCID: PMC6505173
- DOI: 10.1534/g3.119.400165
Cleaning Genotype Data from Diversity Outbred Mice
Abstract
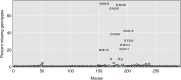
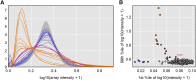
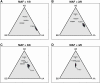
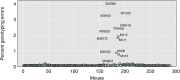
Data cleaning is an important first step in most statistical analyses, including efforts to map the genetic loci that contribute to variation in quantitative traits. Here we illustrate approaches to quality control and cleaning of array-based genotyping data for multiparent populations (experimental crosses derived from more than two founder strains), using MegaMUGA array data from a set of 291 Diversity Outbred (DO) mice. Our approach employs data visualizations that can reveal problems at the level of individual mice or with individual SNP markers. We find that the proportion of missing genotypes for each mouse is an effective indicator of sample quality. We use microarray probe intensities for SNPs on the X and Y chromosomes to confirm the sex of each mouse, and we use the proportion of matching SNP genotypes between pairs of mice to detect sample duplicates. We use a hidden Markov model (HMM) reconstruction of the founder haplotype mosaic across each mouse genome to estimate the number of crossovers and to identify potential genotyping errors. To evaluate marker quality, we find that missing data and genotyping error rates are the most effective diagnostics. We also examine the SNP genotype frequencies with markers grouped according to their minor allele frequency in the founder strains. For markers with high apparent error rates, a scatterplot of the allele-specific probe intensities can reveal the underlying cause of incorrect genotype calls. The decision to include or exclude low-quality samples can have a significant impact on the mapping results for a given study. We find that the impact of low-quality markers on a given study is often minimal, but reporting problematic markers can improve the utility of the genotyping array across many studies.
Keywords: MPP; Multiparent Advanced Generation Inter-Cross (MAGIC); QTL; data cleaning; data diagnostics; multiparental populations; quantitative trait loci.
Copyright © 2019 Broman et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
