DamID as a versatile tool for understanding gene regulation
- PMID: 30877125
- PMCID: PMC6451315
- DOI: 10.1242/dev.173666
DamID as a versatile tool for understanding gene regulation
Abstract
The interaction of proteins and RNA with chromatin underlies the regulation of gene expression. The ability to profile easily these interactions is fundamental for understanding chromatin biology in vivo DNA adenine methyltransferase identification (DamID) profiles genome-wide protein-DNA interactions without antibodies, fixation or protein pull-downs. Recently, DamID has been adapted for applications beyond simple assaying of protein-DNA interactions, such as for studying RNA-chromatin interactions, chromatin accessibility and long-range chromosome interactions. Here, we provide an overview of DamID and introduce improvements to the technology, discuss their applications and compare alternative methodologies.
Keywords: Chromatin; DamID; Gene regulation; Genomics.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
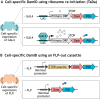
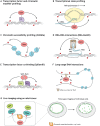
Similar articles
-
DamID as an approach to studying long-distance chromatin interactions.Methods Mol Biol. 2014;1196:279-89. doi: 10.1007/978-1-4939-1242-1_17. Methods Mol Biol. 2014. PMID: 25151170
-
Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase.Nat Biotechnol. 2000 Apr;18(4):424-8. doi: 10.1038/74487. Nat Biotechnol. 2000. PMID: 10748524
-
iDamIDseq and iDEAR: an improved method and computational pipeline to profile chromatin-binding proteins.Development. 2016 Nov 15;143(22):4272-4278. doi: 10.1242/dev.139261. Epub 2016 Oct 5. Development. 2016. PMID: 27707796 Free PMC article.
-
DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase.Methods Enzymol. 2006;410:342-59. doi: 10.1016/S0076-6879(06)10016-6. Methods Enzymol. 2006. PMID: 16938559 Review.
-
DamID: a methylation-based chromatin profiling approach.Methods Mol Biol. 2009;567:155-69. doi: 10.1007/978-1-60327-414-2_11. Methods Mol Biol. 2009. PMID: 19588092 Review.
Cited by
-
Tethered MNase Structure Probing as Versatile Technique for Analyzing RNPs Using Tagging Cassettes for Homologous Recombination in Saccharomyces cerevisiae.Methods Mol Biol. 2022;2533:127-145. doi: 10.1007/978-1-0716-2501-9_8. Methods Mol Biol. 2022. PMID: 35796986 Free PMC article.
-
Host RNA N6-methyladenosine and incoming DNA N6-methyldeoxyadenosine modifications cooperatively elevate the condensation potential of DNA to activate immune surveillance.Mol Ther. 2024 Dec 4;32(12):4418-4434. doi: 10.1016/j.ymthe.2024.10.027. Epub 2024 Oct 28. Mol Ther. 2024. PMID: 39473181 Free PMC article.
-
The role of inner nuclear membrane protein emerin in myogenesis.FASEB J. 2025 Apr 15;39(7):e70514. doi: 10.1096/fj.202500323. FASEB J. 2025. PMID: 40178931 Free PMC article. Review.
-
Transcriptional Repression of reaper by Stand Still Safeguards Female Germline Development in Drosophila.bioRxiv [Preprint]. 2025 Jun 3:2025.05.17.654630. doi: 10.1101/2025.05.17.654630. bioRxiv. 2025. PMID: 40501627 Free PMC article. Preprint.
-
Evolving methodologies and concepts in 4D nucleome research.Curr Opin Cell Biol. 2020 Jun;64:105-111. doi: 10.1016/j.ceb.2020.04.005. Epub 2020 May 27. Curr Opin Cell Biol. 2020. PMID: 32473574 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

