Meiotic chromosomes in motion: a perspective from Mus musculus and Caenorhabditis elegans
- PMID: 30877366
- PMCID: PMC6823321
- DOI: 10.1007/s00412-019-00698-5
Meiotic chromosomes in motion: a perspective from Mus musculus and Caenorhabditis elegans
Abstract
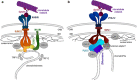
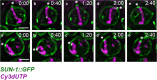
Vigorous chromosome movement during the extended prophase of the first meiotic division is conserved in most eukaryotes. The movement is crucial for the faithful segregation of homologous chromosomes into daughter cells, and thus for fertility. A prerequisite for meiotic chromosome movement is the stable and functional attachment of telomeres or chromosome ends to the nuclear envelope and their cytoplasmic coupling to the cytoskeletal forces responsible for generating movement. Important advances in understanding the components, mechanisms, and regulation of chromosome end attachment and movement have recently been made. This review focuses on insights gained from experiments into two major metazoan model organisms: the mouse, Mus musculus, and the nematode, Caenorhabditis elegans.
Keywords: C. elegans; Chromosome movement; LINC complex; Meiosis; Mouse.
Figures

References
-
- Alsheimer M, Benavente R. Change of karyoskeleton during mammalian spermatogenesis: expression pattern of nuclear lamin C2 and its regulation. Exp Cell Res. 1996;228:181–188. - PubMed
-
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

