Ccne1 Overexpression Causes Chromosome Instability in Liver Cells and Liver Tumor Development in Mice
- PMID: 30878468
- PMCID: PMC6800187
- DOI: 10.1053/j.gastro.2019.03.016
Ccne1 Overexpression Causes Chromosome Instability in Liver Cells and Liver Tumor Development in Mice
Abstract
Background & aims: The CCNE1 locus, which encodes cyclin E1, is amplified in many types of cancer cells and is activated in hepatocellular carcinomas (HCCs) from patients infected with hepatitis B virus or adeno-associated virus type 2, due to integration of the virus nearby. We investigated cell-cycle and oncogenic effects of cyclin E1 overexpression in tissues of mice.
Methods: We generated mice with doxycycline-inducible expression of Ccne1 (Ccne1T mice) and activated overexpression of cyclin E1 from age 3 weeks onward. At 14 months of age, livers were collected from mice that overexpress cyclin E1 and nontransgenic mice (controls) and analyzed for tumor burden and by histology. Mouse embryonic fibroblasts (MEFs) and hepatocytes from Ccne1T and control mice were analyzed to determine the extent to which cyclin E1 overexpression perturbs S-phase entry, DNA replication, and numbers and structures of chromosomes. Tissues from 4-month-old Ccne1T and control mice (at that age were free of tumors) were analyzed for chromosome alterations, to investigate the mechanisms by which cyclin E1 predisposes hepatocytes to transformation.
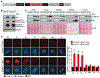
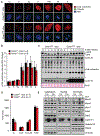
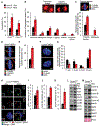
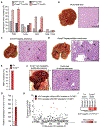
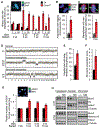
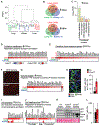
Results: Ccne1T mice developed more hepatocellular adenomas and HCCs than control mice. Tumors developed only in livers of Ccne1T mice, despite high levels of cyclin E1 in other tissues. Ccne1T MEFs had defects that promoted chromosome missegregation and aneuploidy, including incomplete replication of DNA, centrosome amplification, and formation of nonperpendicular mitotic spindles. Whereas Ccne1T mice accumulated near-diploid aneuploid cells in multiple tissues and organs, polyploidization was observed only in hepatocytes, with losses and gains of whole chromosomes, DNA damage, and oxidative stress.
Conclusions: Livers, but not other tissues of mice with inducible overexpression of cyclin E1, develop tumors. More hepatocytes from the cyclin E1-overexpressing mice were polyploid than from control mice, and had losses or gains of whole chromosomes, DNA damage, and oxidative stress; all of these have been observed in human HCC cells. The increased risk of HCC in patients with hepatitis B virus or adeno-associated virus type 2 infection might involve activation of cyclin E1 and its effects on chromosomes and genomes of liver cells.
Keywords: AAV2; Chromosome Integrity; HBV; Hepatocarcinogenesis.
Copyright © 2019 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

