A complete protocol for whole-genome sequencing of virus from clinical samples: Application to coronavirus OC43
- PMID: 30878524
- PMCID: PMC7112119
- DOI: 10.1016/j.virol.2019.03.006
A complete protocol for whole-genome sequencing of virus from clinical samples: Application to coronavirus OC43
Abstract
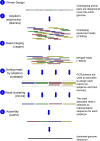
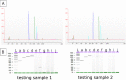
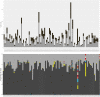
Genome sequencing of virus has become a useful tool for better understanding of virus pathogenicity and epidemiological surveillance. Obtaining virus genome sequence directly from clinical samples is still a challenging task due to the low load of virus genetic material compared to the host DNA, and to the difficulty to get an accurate genome assembly. Here we introduce a complete sequencing and analyzing protocol called V-ASAP for Virus Amplicon Sequencing Assembly Pipeline. Our protocol is able to generate the viral dominant genome sequence starting from clinical samples. It is based on a multiplex PCR amplicon sequencing coupled with a reference-free analytical pipeline. This protocol was applied to 11 clinical samples infected with coronavirus OC43 (HcoV-OC43), and led to seven complete and two nearly complete genome assemblies. The protocol introduced here is shown to be robust, to produce a reliable sequence, and could be applied to other virus.
Keywords: Bioinformatics; Complete genome; Coronavirus; High-throughput sequencing.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Cotten M., Lam T.T., Watson S.J., Palser A.L., Petrova V., Grant P., Pybus O.G., Rambaut A., Guan Y., Pillay D., Kellam P., Nastouli E. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg. Infect. Dis. 2013;19 doi: 10.3201/eid1905.130057. (736–42B) - DOI - PMC - PubMed
-
- Dinwiddie D.L., Hardin O., Denson J.L., Kincaid J.C., Schwalm K.C., Stoner A.N., Abramo T.J., Thompson T.M., Putt C.M., Young S.A., Dehority W.N., Kennedy J.L. Complete genome sequences of four novel human coronavirus OC43 isolates associated with severe acute respiratory infection. Genome Announc. 2018;6 doi: 10.1128/genomeA.00452-18. (e00452-18) - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

