Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients
- PMID: 30878974
- PMCID: PMC6579551
- DOI: 10.1136/annrheumdis-2018-214539
Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients
Abstract
Objectives: To unravel the hierarchy of cellular/molecular pathways in the disease tissue of early, treatment-naïve rheumatoid arthritis (RA) patients and determine their relationship with clinical phenotypes and treatment response/outcomes longitudinally.
Methods: 144 consecutive treatment-naïve early RA patients (<12 months symptoms duration) underwent ultrasound-guided synovial biopsy before and 6 months after disease-modifying antirheumatic drug (DMARD) initiation. Synovial biopsies were analysed for cellular (immunohistology) and molecular (NanoString) characteristics and results compared with clinical and imaging outcomes. Differential gene expression analysis and logistic regression were applied to define variables correlating with treatment response and predicting radiographic progression.
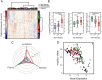
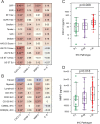
Results: Cellular and molecular analyses of synovial tissue demonstrated for the first time in early RA the presence of three pathology groups: (1) lympho-myeloid dominated by the presence of B cells in addition to myeloid cells; (2) diffuse-myeloid with myeloid lineage predominance but poor in B cells nd (3) pauci-immune characterised by scanty immune cells and prevalent stromal cells. Longitudinal correlation of molecular signatures demonstrated that elevation of myeloid- and lymphoid-associated gene expression strongly correlated with disease activity, acute phase reactants and DMARD response at 6 months. Furthermore, elevation of synovial lymphoid-associated genes correlated with autoantibody positivity and elevation of osteoclast-targeting genes predicting radiographic joint damage progression at 12 months. Patients with predominant pauci-immune pathology showed less severe disease activity and radiographic progression.
Conclusions: We demonstrate at disease presentation, prior to pathology modulation by therapy, the presence of specific cellular/molecular synovial signatures that delineate disease severity/progression and therapeutic response and may pave the way to more precise definition of RA taxonomy, therapeutic targeting and improved outcomes.
Keywords: dmards (synthetic); early rheumatoid arthritis; inflammation; synovitis.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY. Published by BMJ.
Figures




Comment in
-
'Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients'.Ann Rheum Dis. 2020 Nov;79(11):e140. doi: 10.1136/annrheumdis-2019-215881. Epub 2019 Jul 13. Ann Rheum Dis. 2020. PMID: 31302597 No abstract available.
-
Response to: 'Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients' by Buch et al.Ann Rheum Dis. 2020 Nov;79(11):e141. doi: 10.1136/annrheumdis-2019-215903. Epub 2019 Jul 13. Ann Rheum Dis. 2020. PMID: 31302598 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

