Glycolysis Regulates Human Embryonic Stem Cell Self-Renewal under Hypoxia through HIF-2α and the Glycolytic Sensors CTBPs
- PMID: 30880076
- PMCID: PMC6450050
- DOI: 10.1016/j.stemcr.2019.02.005
Glycolysis Regulates Human Embryonic Stem Cell Self-Renewal under Hypoxia through HIF-2α and the Glycolytic Sensors CTBPs
Abstract
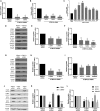
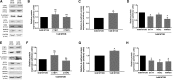
Glycolysis and hypoxia are key regulators of human embryonic stem cell (hESC) self-renewal, but how changes in metabolism affect gene expression is poorly understood. C-terminal binding proteins (CTBPs) are glycolytic sensors that through NADH binding link the metabolic state of the cell to its gene expression, by acting as transcriptional corepressors, or coactivators. However, the role of CTBPs in hESCs has not previously been investigated. A direct interaction between hypoxia-inducible factor 2α (HIF-2α) and the CTBP proximal promoters in hESCs cultured only under hypoxia was demonstrated. Decreasing the rate of flux through glycolysis in hESCs maintained under hypoxia resulted in a reduction of CTBPs, OCT4, SOX2, and NANOG, but also in the expression of HIF-2α. Silencing CTBP expression resulted in the loss of pluripotency marker expression demonstrating that CTBPs are involved in hESC maintenance. These data suggest that under hypoxia, glycolysis regulates self-renewal through HIF-2α and the induction of the metabolic sensors CTBPs.
Keywords: C-terminal binding proteins; NANOG; OCT4; SOX2; glycolysis; human embryonic stem cells; hypoxia-inducible factors; metabolism; self-renewal.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Boyd J.M., Subramanian T., Schaeper U., La Regina M., Bayley S., Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. - PMC - PubMed
-
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. - PubMed
-
- Chen H.F., Kuo H.C., Lin S.P., Chien C.L., Chiang M.S., Ho H.N. Hypoxic culture maintains self-renewal and enhances embryoid body formation of human embryonic stem cells. Tissue Eng. Part A. 2010;16:2901–2913. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

