Tracing MYC Expression for Small Molecule Discovery
- PMID: 30880156
- PMCID: PMC6525070
- DOI: 10.1016/j.chembiol.2019.02.007
Tracing MYC Expression for Small Molecule Discovery
Abstract
Our inability to effectively "drug" targets such as MYC for therapeutic purposes requires the development of new approaches. We report on the implementation of a phenotype-based assay for monitoring MYC expression in multiple myeloma cells. The open reading frame (ORF) encoding an unstable variant of GFP was engineered immediately downstream of the MYC ORF using CRISPR/Cas9, resulting in co-expression of both proteins from the endogenous MYC locus. Using fluorescence readout as a surrogate for MYC expression, we implemented a pilot screen in which ∼10,000 compounds were prosecuted. Among known MYC expression inhibitors, we identified cardiac glycosides and cytoskeletal disruptors to be quite potent. We demonstrate the power of CRISPR/Cas9 engineering in establishing phenotype-based assays to identify gene expression modulators.
Keywords: CRISPR/Cas9; MYC expression; chemical biology; drug discovery; drug repurposing; genome engineering; multiple myelomas; phenotype-based assay; target identification.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Figures

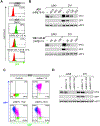

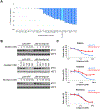


References
-
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, and Topisirovic I (2015). Targeting the translation machinery in cancer. Nat Rev Drug Discov 14, 261–278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

