Antimicrobial resistance, virulence genes profiling and molecular relatedness of methicillin-resistant Staphylococcus aureus strains isolated from hospitalized patients in Guangdong Province, China
- PMID: 30881052
- PMCID: PMC6394240
- DOI: 10.2147/IDR.S192611
Antimicrobial resistance, virulence genes profiling and molecular relatedness of methicillin-resistant Staphylococcus aureus strains isolated from hospitalized patients in Guangdong Province, China
Abstract
Purpose: The main objective of this study was to decipher the prevalence, antimicrobial resistance, major virulence genes and the molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) isolated from different clinical sources in southern China.
Materials and methods: The present study was performed on 187 non-duplicate S. aureus clinical isolates collected from three tertiary hospitals in Guangdong Province, China, 2010-2016. Antimicrobial susceptibility testing was performed by the disk diffusion method and by measuring the minimum inhibitory concentration. Screening for resistance and virulence genes was performed. Clonal relatedness was determined using various molecular typing methods such as multilocus sequence typing, spa and staphylococcal chromosomal cassette mec (SCCmec) typing. Whole genome sequencing was performed for three selected isolates.
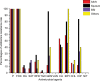
Results: Out of 187 isolates, 103 (55%) were identified as MRSA. The highest prevalence rate was found among the skin and soft tissue infection (SSTI) samples (58/103), followed by sputum samples (25/103), blood stream infection samples (15/103) and others (5/103). Antimicrobial susceptibility results revealed high resistance rates for erythromycin (64.1%), clindamycin (48.5%), gentamicin (36.9%) and ciprofloxacin (33.98%). All isolates were susceptible to vancomycin. Resistance genes and mutation detected were as follows: aac(6')-aph(2") (24.3%), dfrG (10.7%), rpoB (21.4%), cfr (0%), fexA (1.94%), gyrA (35.92%), gyrB (0.97%), grlA (20.4%), grlB (10.68%), ermA (21.4%), ermB (18.44%), ermC (21.4%) and lnuA (18.44%). Profiling of virulence genes revealed the following: sea (11.7%), seb (21.4%), sec (0.97%), sed (0.97%), hla (86.41%), hlb (17.48%), hlg (10.68%), hld (53.4%), Tsst-1 (3.9%) and pvl (27.2%). Clonal relatedness showed that ST239-SCCmecA III-t37 clone was the most prevalent clone.
Conclusion: Our study elucidated the prevalence, antibiotic resistance, pathogenicity and molecular characteristics of MRSA isolated from various clinical sources in Guangdong, China. We found that the infectious rate of MRSA was higher among SSTI than other sources. The most predominant genotype was ST239-SCCmecA III-t37 clone, indicating that ST239-t30 clone which was previously predominant had been replaced by a new clone.
Keywords: MRSA; antibiotic resistance; molecular typing; resistance genes; virulence factors; whole genome sequencing.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures

References
-
- Goudarzi M, Seyedjavadi SS, Nasiri MJ, Goudarzi H, Sajadi Nia R, Dabiri H. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb Pathog. 2017;104:328–335. - PubMed
-
- Jiang B, Yin S, You B, et al. Antimicrobial resistance and virulence genes profiling of methicillin-resistant Staphylococcus aureus isolates in a burn center: a 5-year study. Microb Pathog. 2018;114:176–179. - PubMed
-
- Papadopoulos P, Papadopoulos T, Angelidis AS, et al. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018;69:43–50. - PubMed
-
- Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKA-PEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14(8):529–542. - PubMed
-
- Savoldi A, Azzini AM, Baur D, Tacconelli E. Is there still a role for vancomycin in skin and soft-tissue infections? Curr Opin Infect Dis. 2018;31(2):120–130. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

