Effect of Nucleosome Assembly on Alkylation by a Dynamic Electrophile
- PMID: 30882212
- PMCID: PMC6533127
- DOI: 10.1021/acs.chemrestox.9b00057
Effect of Nucleosome Assembly on Alkylation by a Dynamic Electrophile
Abstract
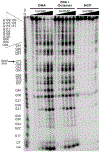
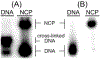
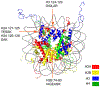
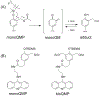
Quinone methides are reactive electrophiles that are generated during metabolism of various drugs, natural products, and food additives. Their chemical properties and cellular effects have been described previously, and now their response to packaging DNA in a nucleosome core is described. A model bisquinone methide precursor (bisQMP) was selected based on its ability to form reversible adducts with guanine N7 that allow for their redistribution and transfer after quinone methide regeneration. Assembly of Widom's 601 DNA with the histone octamer of H2A, H2B, H3, and H4 from Xenopus laevis significantly suppressed alkylation of the DNA. This result is a function of DNA packaging since addition of the octamer without nucleosome reconstitution only mildly protected DNA from alkylation. The lack of competition between nucleophiles of DNA and the histones was consistent with the limited number of adducts formed by the histones as detected by tryptic digestion and ultraperformance liquid chromatography-mass spectrometry. Only three peptide adducts were observed after reaction with a monofunctional analogue of bisQMP, and only two peptide adducts were observed after reaction with bisQMP. Histone reaction was also suppressed when reconstituted into the nucleosome core particle. However, bisQMP was capable of cross-linking the DNA and histones in moderate yields (∼20%) that exceeded expectations derived from reaction of cisplatin, nitrogen mustards, and diepoxybutane. The core histones also demonstrated a protective function against dynamic alkylation by trapping the reactive quinone methide after its spontaneous regeneration from DNA adducts.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Directing Quinone Methide-Dependent Alkylation and Cross-Linking of Nucleic Acids with Quaternary Amines.Bioconjug Chem. 2020 May 20;31(5):1486-1496. doi: 10.1021/acs.bioconjchem.0c00166. Epub 2020 Apr 23. Bioconjug Chem. 2020. PMID: 32298588 Free PMC article.
-
Association of nucleosome core particle DNA with different histone oligomers. Transfer of histones between DNA-(H2A,H2B) and DNA-(H3,H4) complexes.J Mol Biol. 1988 Nov 5;204(1):141-54. doi: 10.1016/0022-2836(88)90605-5. J Mol Biol. 1988. PMID: 3216389
-
Asymmetric linker histone association directs the asymmetric rearrangement of core histone interactions in a positioned nucleosome containing a thyroid hormone response element.Biochemistry. 1998 Jun 16;37(24):8629-36. doi: 10.1021/bi9805846. Biochemistry. 1998. PMID: 9628724
-
Quinone methide derivatives: important intermediates to DNA alkylating and DNA cross-linking actions.Curr Med Chem. 2005;12(24):2893-913. doi: 10.2174/092986705774454724. Curr Med Chem. 2005. PMID: 16305478 Review.
-
Chromatin structures condensed by linker histones.Essays Biochem. 2019 Apr 23;63(1):75-87. doi: 10.1042/EBC20180056. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 31015384 Review.
Cited by
-
Unraveling Reversible DNA Cross-Links with a Biological Machine.Chem Res Toxicol. 2020 Nov 16;33(11):2903-2913. doi: 10.1021/acs.chemrestox.0c00413. Epub 2020 Nov 5. Chem Res Toxicol. 2020. PMID: 33147957 Free PMC article.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. - PubMed
-
- Millard JT (1996) DNA modifying agents as tools for studying chromatin structure. Biochimie 78, 803–816. - PubMed
-
- Millard JT, Spencer RJ, and Hopkins PB (1998) Effect of nucleosome structure on DNA interstrand cross-linking reactions. Biochemistry 37, 5211–5219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

