Role of the global regulator Rex in control of NAD+ -regeneration in Clostridioides (Clostridium) difficile
- PMID: 30882947
- PMCID: PMC6561804
- DOI: 10.1111/mmi.14245
Role of the global regulator Rex in control of NAD+ -regeneration in Clostridioides (Clostridium) difficile
Abstract
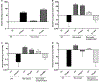
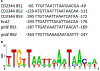
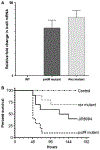
For the human pathogen Clostridioides (also known as Clostridium) difficile, the ability to adapt to nutrient availability is critical for its proliferation and production of toxins during infection. Synthesis of the toxins is regulated by the availability of certain carbon sources, fermentation products and amino acids (e.g. proline, cysteine, isoleucine, leucine and valine). The effect of proline is attributable at least in part to its role as an inducer and substrate of D-proline reductase (PR), a Stickland reaction that regenerates NAD+ from NADH. Many Clostridium spp. use Stickland metabolism (co-fermentation of pairs of amino acids) to generate ATP and NAD+ . Synthesis of PR is activated by PrdR, a proline-responsive regulatory protein. Here we report that PrdR, in the presence of proline, represses other NAD+ -generating pathways, such as the glycine reductase and succinate-acetyl CoA utilization pathways leading to butyrate production, but does so indirectly by affecting the activity of Rex, a global redox-sensing regulator that responds to the NAD+ /NADH ratio. Our results indicate that PR activity is the favored mechanism for NAD+ regeneration and that both Rex and PrdR influence toxin production. Using the hamster model of C. difficile infection, we revealed the importance of PrdR-regulated Stickland metabolism in the virulence of C. difficile.
© 2019 John Wiley & Sons Ltd.
Figures







References
-
- Antunes A, Martin-Verstraete I & Dupuy B, (2011) CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79: 882–899. - PubMed
-
- Bailey TL & Elkan C, (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. - PubMed
-
- Battaglioli EJ, Hale VL, Chen J, Jeraldo P, Ruiz-Mojica C, Schmidt BA, Rekdal VM, Till LM, Huq L, Smits SA, Moor WJ, Jones-Hall Y, Smyrk T, Khanna S, Pardi DS, Grover M, Patel R, Chia N, Nelson H, Sonnenburg JL, Farrugia G & Kashyap PC, (2018) Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Science translational medicine 10. - PMC - PubMed
-
- Benjamini Y & Hochberg Y, (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 57: 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

