Dopamine D4 Receptor-Selective Compounds Reveal Structure-Activity Relationships that Engender Agonist Efficacy
- PMID: 30883109
- PMCID: PMC6466480
- DOI: 10.1021/acs.jmedchem.9b00231
Dopamine D4 Receptor-Selective Compounds Reveal Structure-Activity Relationships that Engender Agonist Efficacy
Abstract
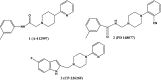
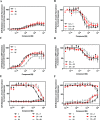
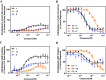
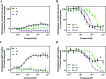
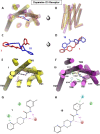
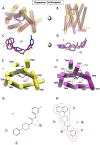
The dopamine D4 receptor (D4R) plays important roles in cognition, attention, and decision making. Novel D4R-selective ligands have promise in medication development for neuropsychiatric conditions, including Alzheimer's disease and substance use disorders. To identify new D4R-selective ligands, and to understand the molecular determinants of agonist efficacy at D4R, we report a series of eighteen novel ligands based on the classical D4R agonist A-412997 (1, 2-(4-(pyridin-2-yl)piperidin-1-yl)- N-( m-tolyl)acetamide). Compounds were profiled using radioligand binding displacement assays, β-arrestin recruitment assays, cyclic AMP inhibition assays, and molecular dynamics computational modeling. We identified several novel D4R-selective ( Ki ≤ 4.3 nM and >100-fold vs other D2-like receptors) compounds with diverse partial agonist and antagonist profiles, falling into three structural groups. These compounds highlight receptor-ligand interactions that control efficacy at D2-like receptors and may provide insights into targeted drug discovery, leading to a better understanding of the role of D4Rs in neuropsychiatric disorders.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- González S.; Rangel-Barajas C.; Peper M.; Lorenzo R.; Moreno E.; Ciruela F.; Borycz J.; Ortiz J.; Lluís C.; Franco R.; McCormick P. J.; Volkow N. D.; Rubinstein M.; Floran B.; Ferré S. Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain. Mol. Psychiatr. 2012, 17, 650–662. 10.1038/mp.2011.93. - DOI - PMC - PubMed
-
- Woolley M. L.; Waters K. A.; Reavill C.; Bull S.; Lacroix L. P.; Martyn A. J.; Hutcheson D. M.; Valerio E.; Bate S.; Jones D. N. C.; Dawson L. A. Selective dopamine D4 receptor agonist (A-412997) improves cognitive performance and stimulates motor activity without influencing reward-related behaviour in rat. Behav. Pharmacol. 2008, 19, 765–776. 10.1097/fbp.0b013e32831c3b06. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

