Activity-Based DNA-Encoded Library Screening
- PMID: 30884226
- PMCID: PMC6786493
- DOI: 10.1021/acscombsci.9b00037
Activity-Based DNA-Encoded Library Screening
Abstract
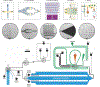
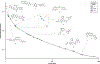
Robotic high-throughput compound screening (HTS) and, increasingly, DNA-encoded library (DEL) screening are driving bioactive chemical matter discovery in the postgenomic era. HTS enables activity-based investigation of highly complex targets using static compound libraries. Conversely, DEL grants efficient access to novel chemical diversity, although screening is limited to affinity-based selections. Here, we describe an integrated droplet-based microfluidic circuit that directly screens solid-phase DELs for activity. An example screen of a 67 100-member library for inhibitors of the phosphodiesterase autotaxin yielded 35 high-priority structures for nanomole-scale synthesis and validation (20 active), guiding candidate selection for synthesis at scale (5/5 compounds with IC50 values of 4-10 μM). We further compared activity-based hits with those of an analogous affinity-based DEL selection. This miniaturized screening platform paves the way toward applying DELs to more complex targets (signaling pathways, cellular response) and represents a distributable approach to small molecule discovery.
Keywords: DNA-encoded library; OBOC; droplet; drug discovery; microfluidics.
Figures



References
-
- Brown DG; Boström J Where Do Recent Small Molecule Clinical Development Candidates Come From? J Med Chem 2018, 61, 9442–9468. - PubMed
-
- Scott DE; Bayly AR; Abell C; Skidmore J Small Molecules, Big Targets: Drug Discovery Faces the Protein-Protein Interaction Challenge. Nat Rev Drug Discov 2016, 15, 533–550. - PubMed
-
- Tommasi R; Brown DG; Walkup GK; Manchester JI; Miller AA ESKAPEing the Labyrinth of Antibacterial Discovery. Nat Rev Drug Discov 2015, 14, 529–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

