Inhibition of Triple-Negative Breast Cancer Cell Aggressiveness by Cathepsin D Blockage: Role of Annexin A1
- PMID: 30884823
- PMCID: PMC6471925
- DOI: 10.3390/ijms20061337
Inhibition of Triple-Negative Breast Cancer Cell Aggressiveness by Cathepsin D Blockage: Role of Annexin A1
Abstract
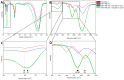
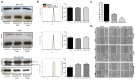
Triple-negative breast cancers (TNBCs) are more aggressive than other breast cancer (BC) subtypes and lack effective therapeutic options. Unraveling marker events of TNBCs may provide new directions for development of strategies for targeted TNBC therapy. Herein, we reported that Annexin A1 (AnxA1) and Cathepsin D (CatD) are highly expressed in MDA-MB-231 (TNBC lineage), compared to MCF-10A and MCF-7. Since the proposed concept was that CatD has protumorigenic activity associated with its ability to cleave AnxA1 (generating a 35.5 KDa fragment), we investigated this mechanism more deeply using the inhibitor of CatD, Pepstatin A (PepA). Fourier Transform Infrared (FTIR) spectroscopy demonstrated that PepA inhibits CatD activity by occupying its active site; the OH bond from PepA interacts with a CO bond from carboxylic acids of CatD catalytic aspartate dyad, favoring the deprotonation of Asp33 and consequently inhibiting CatD. Treatment of MDA-MB-231 cells with PepA induced apoptosis and autophagy processes while reducing the proliferation, invasion, and migration. Finally, in silico molecular docking demonstrated that the catalytic inhibition comprises Asp231 protonated and Asp33 deprotonated, proving all functional results obtained. Our findings elucidated critical CatD activity in TNBC cell trough AnxA1 cleavage, indicating the inhibition of CatD as a possible strategy for TNBC treatment.
Keywords: Annexin A1; Cathepsin D; protease inhibition; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

