Fate of CMY-2-Encoding Plasmids Introduced into the Human Fecal Microbiota by Exogenous Escherichia coli
- PMID: 30885897
- PMCID: PMC6496067
- DOI: 10.1128/AAC.02528-18
Fate of CMY-2-Encoding Plasmids Introduced into the Human Fecal Microbiota by Exogenous Escherichia coli
Abstract
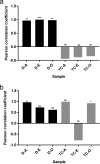
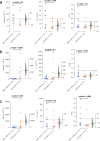
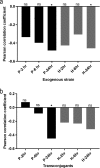
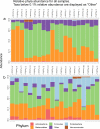
The gut is a hot spot for transfer of antibiotic resistance genes from ingested exogenous bacteria to the indigenous microbiota. The objective of this study was to determine the fate of two nearly identical blaCMY-2-harboring plasmids introduced into the human fecal microbiota by two Escherichia coli strains isolated from a human and from poultry meat. The chromosome and the CMY-2-encoding plasmid of both strains were labeled with distinct fluorescent markers (mCherry and green fluorescent protein [GFP]), allowing fluorescence-activated cell sorting (FACS)-based tracking of the strain and the resident bacteria that have acquired its plasmid. Each strain was introduced into an established in vitro gut model (CoMiniGut) inoculated with individual feces from ten healthy volunteers. Fecal samples collected 2, 6, and 24 h after strain inoculation were analyzed by FACS and plate counts. Although the human strain survived better than the poultry meat strain, both strains transferred their plasmids to the fecal microbiota at concentrations as low as 102 CFU/ml. Strain survival and plasmid transfer varied significantly depending on inoculum concentration and individual fecal microbiota. Identification of transconjugants by 16S rRNA gene sequencing and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) revealed that the plasmids were predominantly acquired by Enterobacteriaceae species, such as E. coli and Hafnia alvei Our experimental data demonstrate that exogenous E. coli of human or animal origin can readily transfer CMY-2-encoding IncI1 plasmids to the human fecal microbiota. Small amounts of the exogenous strain are sufficient to ensure plasmid transfer if the strain is able to survive the gastric environment.
Keywords: CoMiniGut model; Escherichia coli; IncI1; cephalosporin.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Host-Specific Patterns of Genetic Diversity among IncI1-Iγ and IncK Plasmids Encoding CMY-2 β-Lactamase in Escherichia coli Isolates from Humans, Poultry Meat, Poultry, and Dogs in Denmark.Appl Environ Microbiol. 2016 Jul 15;82(15):4705-14. doi: 10.1128/AEM.00495-16. Print 2016 Aug 1. Appl Environ Microbiol. 2016. PMID: 27235431 Free PMC article.
-
A culture-independent method for studying transfer of IncI1 plasmids from wild-type Escherichia coli in complex microbial communities.J Microbiol Methods. 2018 Sep;152:18-26. doi: 10.1016/j.mimet.2018.07.009. Epub 2018 Jul 18. J Microbiol Methods. 2018. PMID: 30030013
-
Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm.Int J Food Microbiol. 2018 Mar 23;269:60-63. doi: 10.1016/j.ijfoodmicro.2018.01.017. Epub 2018 Jan 31. Int J Food Microbiol. 2018. PMID: 29421359
-
Expansive spread of IncI1 plasmids carrying blaCMY-2 amongst Escherichia coli.Int J Antimicrob Agents. 2014 Sep;44(3):203-8. doi: 10.1016/j.ijantimicag.2014.04.016. Epub 2014 Jun 6. Int J Antimicrob Agents. 2014. PMID: 25052868
-
Competition between Escherichia coli Populations with and without Plasmids Carrying a Gene Encoding Extended-Spectrum Beta-Lactamase in the Broiler Chicken Gut.Appl Environ Microbiol. 2019 Aug 14;85(17):e00892-19. doi: 10.1128/AEM.00892-19. Print 2019 Sep 1. Appl Environ Microbiol. 2019. PMID: 31253677 Free PMC article.
Cited by
-
High-resolution characterisation of ESBL/pAmpC-producing Escherichia coli isolated from the broiler production pyramid.Sci Rep. 2020 Jul 7;10(1):11123. doi: 10.1038/s41598-020-68036-9. Sci Rep. 2020. PMID: 32636426 Free PMC article.
-
Drosophila Model for Gut-Mediated Horizontal Transfer of Narrow- and Broad-Host-Range Plasmids.mSphere. 2021 Oct 27;6(5):e0069821. doi: 10.1128/mSphere.00698-21. Epub 2021 Oct 20. mSphere. 2021. PMID: 34668756 Free PMC article.
-
Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed.Antibiotics (Basel). 2023 Mar 14;12(3):569. doi: 10.3390/antibiotics12030569. Antibiotics (Basel). 2023. PMID: 36978436 Free PMC article.
-
Succession in the caecal microbiota of developing broilers colonised by extended-spectrum β-lactamase-producing Escherichia coli.Anim Microbiome. 2022 Aug 19;4(1):51. doi: 10.1186/s42523-022-00199-4. Anim Microbiome. 2022. PMID: 35986389 Free PMC article.
-
Genome-Based Analysis of Extended-Spectrum β-Lactamase-Producing Escherichia coli in the Aquatic Environment and Nile Perch (Lates niloticus) of Lake Victoria, Tanzania.Front Microbiol. 2020 Feb 21;11:108. doi: 10.3389/fmicb.2020.00108. eCollection 2020. Front Microbiol. 2020. PMID: 32153519 Free PMC article.
References
-
- European Centre for Disease Prevention and Control (ECDC), European Food Safety Authority (EFSA), European Medicines Agency (EMA). 2017. ECDC/EFSA/EMA first joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFSA J 15:4872. doi: 10.2903/j.efsa.2017.4872. - DOI - PMC - PubMed
-
- Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schonning K, Agerso Y, Guardabassi L. 2016. Host-specific patterns of genetic diversity among IncI1-I gamma and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 82:4705–4714. doi: 10.1128/AEM.00495-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

