The RNase YbeY Is Vital for Ribosome Maturation, Stress Resistance, and Virulence of the Natural Genetic Engineer Agrobacterium tumefaciens
- PMID: 30885931
- PMCID: PMC6509659
- DOI: 10.1128/JB.00730-18
The RNase YbeY Is Vital for Ribosome Maturation, Stress Resistance, and Virulence of the Natural Genetic Engineer Agrobacterium tumefaciens
Abstract
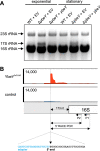
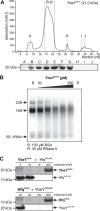
Riboregulation involving regulatory RNAs, RNA chaperones, and ribonucleases is fundamental for the rapid adaptation of gene expression to changing environmental conditions. The gene coding for the RNase YbeY belongs to the minimal prokaryotic genome set and has a profound impact on physiology in a wide range of bacteria. Here, we show that the Agrobacterium tumefaciensybeY gene is not essential. Deletion of the gene in the plant pathogen reduced growth, motility, and stress tolerance. Most interestingly, YbeY is crucial for A. tumefaciens-mediated T-DNA transfer and tumor formation. Comparative proteomics by using isobaric tags for relative and absolute quantitation (iTRAQ) revealed dysregulation of 59 proteins, many of which have previously been found to be dependent on the RNA chaperone Hfq. YbeY and Hfq have opposing effects on production of these proteins. Accumulation of a 16S rRNA precursor in the ybeY mutant suggests that A. tumefaciens YbeY is involved in rRNA processing. RNA coimmunoprecipitation-sequencing (RIP-Seq) showed binding of YbeY to the region immediately upstream of the 16S rRNA. Purified YbeY is an oligomer with RNase activity. It does not physically interact with Hfq and thus plays a partially overlapping but distinct role in the riboregulatory network of the plant pathogen.IMPORTANCE Although ybeY gene belongs to the universal bacterial core genome, its biological function is incompletely understood. Here, we show that YbeY is critical for fitness and host-microbe interaction in the plant pathogen Agrobacterium tumefaciens Consistent with the reported endoribonuclease activity of YbeY, A. tumefaciens YbeY acts as a RNase involved in maturation of 16S rRNA. This report adds a worldwide plant pathogen and natural genetic engineer of plants to the growing list of bacteria that require the conserved YbeY protein for host-microbe interaction.
Keywords: Agrobacterium tumefaciens; Hfq; RNase; YbeY; iTRAQ; rRNA; ribosome; virulence.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Identification of YbeY-Protein Interactions Involved in 16S rRNA Maturation and Stress Regulation in Escherichia coli.mBio. 2016 Nov 8;7(6):e01785-16. doi: 10.1128/mBio.01785-16. mBio. 2016. PMID: 27834201 Free PMC article.
-
Elevated Levels of Era GTPase Improve Growth, 16S rRNA Processing, and 70S Ribosome Assembly of Escherichia coli Lacking Highly Conserved Multifunctional YbeY Endoribonuclease.J Bacteriol. 2018 Aug 10;200(17):e00278-18. doi: 10.1128/JB.00278-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29914987 Free PMC article.
-
Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli.J Biol Chem. 2018 Jun 8;293(23):9006-9016. doi: 10.1074/jbc.RA117.000300. Epub 2018 Apr 20. J Biol Chem. 2018. PMID: 29678883 Free PMC article.
-
The role of the ubiquitin-proteasome system in Agrobacterium tumefaciens-mediated genetic transformation of plants.Plant Physiol. 2012 Sep;160(1):65-71. doi: 10.1104/pp.112.200949. Epub 2012 Jul 10. Plant Physiol. 2012. PMID: 22786890 Free PMC article. Review. No abstract available.
-
Transcriptome Profiling of Plant Genes in Response to Agrobacterium tumefaciens-Mediated Transformation.Curr Top Microbiol Immunol. 2018;418:319-348. doi: 10.1007/82_2018_115. Curr Top Microbiol Immunol. 2018. PMID: 30062593 Review.
Cited by
-
YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa.Appl Environ Microbiol. 2021 Mar 1;87(5):e02171-20. doi: 10.1128/AEM.02171-20. Epub 2020 Dec 11. Appl Environ Microbiol. 2021. PMID: 33310711 Free PMC article.
-
Sinorhizobium meliloti YbeY is a zinc-dependent single-strand specific endoribonuclease that plays an important role in 16S ribosomal RNA processing.Nucleic Acids Res. 2020 Jan 10;48(1):332-348. doi: 10.1093/nar/gkz1095. Nucleic Acids Res. 2020. PMID: 31777930 Free PMC article.
-
Two redox-responsive LysR-type transcription factors control the oxidative stress response of Agrobacterium tumefaciens.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf267. doi: 10.1093/nar/gkaf267. Nucleic Acids Res. 2025. PMID: 40193708 Free PMC article.
-
YBEY is an essential biogenesis factor for mitochondrial ribosomes.Nucleic Acids Res. 2020 Sep 25;48(17):9762-9786. doi: 10.1093/nar/gkaa148. Nucleic Acids Res. 2020. PMID: 32182356 Free PMC article.
References
-
- Mallory AC, Hinze A, Tucker MR, Bouche N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. 2009. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet 5:e1000646. doi:10.1371/journal.pgen.1000646. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

