Inhibition of NF-κB-Dependent Signaling Enhances Sensitivity and Overcomes Resistance to BET Inhibition in Uveal Melanoma
- PMID: 30885979
- PMCID: PMC6643281
- DOI: 10.1158/0008-5472.CAN-18-3177
Inhibition of NF-κB-Dependent Signaling Enhances Sensitivity and Overcomes Resistance to BET Inhibition in Uveal Melanoma
Erratum in
-
Correction: Inhibition of NF-κB-Dependent Signaling Enhances Sensitivity and Overcomes Resistance to BET Inhibition in Uveal Melanoma.Cancer Res. 2025 Aug 1;85(15):2954. doi: 10.1158/0008-5472.CAN-25-2634. Cancer Res. 2025. PMID: 40746072 No abstract available.
Abstract
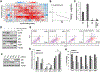
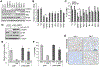
Bromodomain and extraterminal protein inhibitors (BETi) are epigenetic therapies aimed to target dysregulated gene expression in cancer cells. Despite early successes of BETi in a range of malignancies, the development of drug resistance may limit their clinical application. Here, we evaluated the mechanisms of BETi resistance in uveal melanoma, a disease with little treatment options, using two approaches: a high-throughput combinatorial drug screen with the clinical BET inhibitor PLX51107 and RNA sequencing of BETi-resistant cells. NF-κB inhibitors synergistically sensitized uveal melanoma cells to PLX51107 treatment. Furthermore, genes involved in NF-κB signaling were upregulated in BETi-resistant cells, and the transcription factor CEBPD contributed to the mechanism of resistance. These findings suggest that inhibitors of NF-κB signaling may improve the efficacy of BET inhibition in patients with advanced uveal melanoma. SIGNIFICANCE: These findings provide evidence that inhibitors of NF-κB signaling synergize with BET inhibition in in vitro and in vivo models, suggesting a clinical utility of these targeted therapies in patients with uveal melanoma.
©2019 American Association for Cancer Research.
Figures




Similar articles
-
YAP Upregulation Contributes to Acquired Resistance to BET Inhibitors in Uveal Melanoma.Pigment Cell Melanoma Res. 2025 Jul;38(4):e70036. doi: 10.1111/pcmr.70036. Pigment Cell Melanoma Res. 2025. PMID: 40624770
-
Targeting the VIP-VPAC Pathway in Melanoma Models Inhibits Tumor Growth and Liver Metastasis.Cancer Lett. 2025 Sep 28;628:217855. doi: 10.1016/j.canlet.2025.217855. Epub 2025 Jun 5. Cancer Lett. 2025. PMID: 40482913
-
Protein phosphatase 6 activates NF-κB to confer sensitivity to MAPK pathway inhibitors in KRAS- and BRAF-mutant cancer cells.Sci Signal. 2024 May 14;17(836):eadd5073. doi: 10.1126/scisignal.add5073. Epub 2024 May 14. Sci Signal. 2024. PMID: 38743809 Free PMC article.
-
Nanotechnology in Uveal Melanoma: Cutting-Edge Advances to Halt Progression and Metastasis.AAPS PharmSciTech. 2025 Jul 14;26(6):191. doi: 10.1208/s12249-025-03184-7. AAPS PharmSciTech. 2025. PMID: 40660019 Review.
-
Immune checkpoint blockade for unresectable or metastatic uveal melanoma: A systematic review.Cancer Treat Rev. 2017 Nov;60:44-52. doi: 10.1016/j.ctrv.2017.08.009. Epub 2017 Aug 24. Cancer Treat Rev. 2017. PMID: 28881222
Cited by
-
Enhancer rewiring in tumors: an opportunity for therapeutic intervention.Oncogene. 2021 May;40(20):3475-3491. doi: 10.1038/s41388-021-01793-7. Epub 2021 May 1. Oncogene. 2021. PMID: 33934105 Review.
-
The Potential of Antibody Technology and Silver Nanoparticles for Enhancing Photodynamic Therapy for Melanoma.Biomedicines. 2022 Sep 1;10(9):2158. doi: 10.3390/biomedicines10092158. Biomedicines. 2022. PMID: 36140259 Free PMC article. Review.
-
The multi-CDK inhibitor dinaciclib reverses bromo- and extra-terminal domain (BET) inhibitor resistance in acute myeloid leukemia via inhibition of Wnt/β-catenin signaling.Exp Hematol Oncol. 2024 Mar 4;13(1):27. doi: 10.1186/s40164-024-00483-w. Exp Hematol Oncol. 2024. PMID: 38438856 Free PMC article.
-
Mouse models of uveal melanoma: Strengths, weaknesses, and future directions.Pigment Cell Melanoma Res. 2020 Mar;33(2):264-278. doi: 10.1111/pcmr.12853. Epub 2020 Jan 22. Pigment Cell Melanoma Res. 2020. PMID: 31880399 Free PMC article. Review.
-
Emerging Therapeutic Opportunities Based on Current Knowledge of Uveal Melanoma Biology.Cancers (Basel). 2019 Jul 20;11(7):1019. doi: 10.3390/cancers11071019. Cancers (Basel). 2019. PMID: 31330784 Free PMC article. Review.
References
-
- Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, Earle JD, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch Ophthalmol 2005;123: 1639–43. - PubMed
-
- Kujala E, Makitie T, Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci 2003;44: 4651–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

