Low genetic variation is associated with low mutation rate in the giant duckweed
- PMID: 30886148
- PMCID: PMC6423293
- DOI: 10.1038/s41467-019-09235-5
Low genetic variation is associated with low mutation rate in the giant duckweed
Erratum in
-
Publisher Correction: Low genetic variation is associated with low mutation rate in the giant duckweed.Nat Commun. 2019 Apr 16;10(1):1857. doi: 10.1038/s41467-019-09796-5. Nat Commun. 2019. PMID: 30992439 Free PMC article.
Abstract
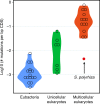
Mutation rate and effective population size (Ne) jointly determine intraspecific genetic diversity, but the role of mutation rate is often ignored. Here we investigate genetic diversity, spontaneous mutation rate and Ne in the giant duckweed (Spirodela polyrhiza). Despite its large census population size, whole-genome sequencing of 68 globally sampled individuals reveals extremely low intraspecific genetic diversity. Assessed under natural conditions, the genome-wide spontaneous mutation rate is at least seven times lower than estimates made for other multicellular eukaryotes, whereas Ne is large. These results demonstrate that low genetic diversity can be associated with large-Ne species, where selection can reduce mutation rates to very low levels. This study also highlights that accurate estimates of mutation rate can help to explain seemingly unexpected patterns of genome-wide variation.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Genome-wide association study of metabolic traits in the giant duckweed Spirodela polyrhiza.Plant Biol (Stuttg). 2025 Jan;27(1):18-28. doi: 10.1111/plb.13747. Epub 2024 Dec 4. Plant Biol (Stuttg). 2025. PMID: 39630110 Free PMC article.
-
Population genomics of the facultatively asexual duckweed Spirodela polyrhiza.New Phytol. 2019 Nov;224(3):1361-1371. doi: 10.1111/nph.16056. Epub 2019 Aug 7. New Phytol. 2019. PMID: 31298732
-
Estimation of the SNP Mutation Rate in Two Vegetatively Propagating Species of Duckweed.G3 (Bethesda). 2020 Nov 5;10(11):4191-4200. doi: 10.1534/g3.120.401704. G3 (Bethesda). 2020. PMID: 32973000 Free PMC article.
-
Status of duckweed genomics and transcriptomics.Plant Biol (Stuttg). 2015 Jan;17 Suppl 1:10-5. doi: 10.1111/plb.12201. Epub 2014 Jul 4. Plant Biol (Stuttg). 2015. PMID: 24995947 Review.
-
Duckweed: Beyond an Efficient Plant Model System.Biomolecules. 2024 May 27;14(6):628. doi: 10.3390/biom14060628. Biomolecules. 2024. PMID: 38927032 Free PMC article. Review.
Cited by
-
Evolution of Mutation Rate in Astronomically Large Phytoplankton Populations.Genome Biol Evol. 2020 Jul 1;12(7):1051-1059. doi: 10.1093/gbe/evaa131. Genome Biol Evol. 2020. PMID: 32645145 Free PMC article.
-
Stability across the Whole Nuclear Genome in the Presence and Absence of DNA Mismatch Repair.Cells. 2021 May 17;10(5):1224. doi: 10.3390/cells10051224. Cells. 2021. PMID: 34067668 Free PMC article. Review.
-
The evolution of the duckweed ionome mirrors losses in structural complexity.Ann Bot. 2024 May 13;133(7):997-1006. doi: 10.1093/aob/mcae012. Ann Bot. 2024. PMID: 38307008 Free PMC article.
-
DNA barcoding and biomass accumulation rates of native Iranian duckweed species for biotechnological applications.Front Plant Sci. 2022 Nov 29;13:1034238. doi: 10.3389/fpls.2022.1034238. eCollection 2022. Front Plant Sci. 2022. PMID: 36523621 Free PMC article.
-
Assembled and annotated 26.5 Gbp coast redwood genome: a resource for estimating evolutionary adaptive potential and investigating hexaploid origin.G3 (Bethesda). 2022 Jan 4;12(1):jkab380. doi: 10.1093/g3journal/jkab380. G3 (Bethesda). 2022. PMID: 35100403 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

