Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output
- PMID: 30886407
- PMCID: PMC6436729
- DOI: 10.1038/s41593-019-0350-2
Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output
Abstract
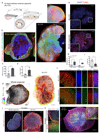
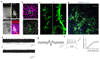
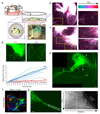
Neural organoids have the potential to improve our understanding of human brain development and neurological disorders. However, it remains to be seen whether these tissues can model circuit formation with functional neuronal output. Here we have adapted air-liquid interface culture to cerebral organoids, leading to improved neuronal survival and axon outgrowth. The resulting thick axon tracts display various morphologies, including long-range projection within and away from the organoid, growth-cone turning, and decussation. Single-cell RNA sequencing reveals various cortical neuronal identities, and retrograde tracing demonstrates tract morphologies that match proper molecular identities. These cultures exhibit active neuronal networks, and subcortical projecting tracts can innervate mouse spinal cord explants and evoke contractions of adjacent muscle in a manner dependent on intact organoid-derived innervating tracts. Overall, these results reveal a remarkable self-organization of corticofugal and callosal tracts with a functional output, providing new opportunities to examine relevant aspects of human CNS development and disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

