Structure-Based Peptide Inhibitor Design of Amyloid-β Aggregation
- PMID: 30886570
- PMCID: PMC6409328
- DOI: 10.3389/fnmol.2019.00054
Structure-Based Peptide Inhibitor Design of Amyloid-β Aggregation
Abstract
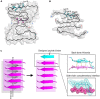
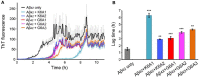
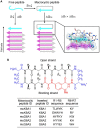
Many human neurodegenerative diseases are associated with amyloid fibril formation. Inhibition of amyloid formation is of importance for therapeutics of the related diseases. However, the development of selective potent amyloid inhibitors remains challenging. Here based on the structures of amyloid β (Aβ) fibrils and their amyloid-forming segments, we designed a series of peptide inhibitors using RosettaDesign. We further utilized a chemical scaffold to constrain the designed peptides into β-strand conformation, which significantly improves the potency of the inhibitors against Aβ aggregation and toxicity. Furthermore, we show that by targeting different Aβ segments, the designed peptide inhibitors can selectively recognize different species of Aβ. Our study developed an approach that combines the structure-based rational design with chemical modification for the development of amyloid inhibitors, which could be applied to the development of therapeutics for different amyloid-related diseases.
Keywords: Alzheimer’s disease; Aβ fibril; neurodegenerative diseases; protein misfolding; structure-based inhibitor design.
Figures


Similar articles
-
Peptide and protein mimetics inhibiting amyloid beta-peptide aggregation.Acc Chem Res. 2008 Oct;41(10):1309-18. doi: 10.1021/ar8000475. Acc Chem Res. 2008. PMID: 18937396
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
-
Assembling amyloid fibrils from designed structures containing a significant amyloid beta-peptide fragment.Biochem J. 2002 Aug 15;366(Pt 1):343-51. doi: 10.1042/BJ20020229. Biochem J. 2002. PMID: 12023906 Free PMC article.
-
Peptide-based amyloid-beta aggregation inhibitors.RSC Med Chem. 2024 Dec 31. doi: 10.1039/d4md00729h. Online ahead of print. RSC Med Chem. 2024. PMID: 39882170 Free PMC article. Review.
-
Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange.Acc Chem Res. 2010 Aug 17;43(8):1072-9. doi: 10.1021/ar9002784. Acc Chem Res. 2010. PMID: 20557067
Cited by
-
Heptameric Peptide Interferes with Amyloid-β Aggregation by Structural Reorganization of the Toxic Oligomers.ACS Omega. 2020 Jun 25;5(26):16128-16138. doi: 10.1021/acsomega.0c01730. eCollection 2020 Jul 7. ACS Omega. 2020. PMID: 32656435 Free PMC article.
-
A Snake Venom Peptide and Its Derivatives Prevent Aβ42 Aggregation and Eliminate Toxic Aβ42 Aggregates In Vitro.ACS Chem Neurosci. 2024 Jul 17;15(14):2600-2611. doi: 10.1021/acschemneuro.4c00089. Epub 2024 Jul 3. ACS Chem Neurosci. 2024. PMID: 38957957 Free PMC article.
-
High-throughput screening for amyloid-β binding natural small-molecules based on the combinational use of biolayer interferometry and UHPLC-DAD-Q/TOF-MS/MS.Acta Pharm Sin B. 2022 Apr;12(4):1723-1739. doi: 10.1016/j.apsb.2021.08.030. Epub 2021 Sep 4. Acta Pharm Sin B. 2022. PMID: 35847494 Free PMC article.
-
Kinetic Control of Parallel versus Antiparallel Amyloid Aggregation via Shape of the Growing Aggregate.Sci Rep. 2019 Nov 5;9(1):15987. doi: 10.1038/s41598-019-52238-x. Sci Rep. 2019. PMID: 31690748 Free PMC article.
-
Substoichiometric Inhibition of Insulin against IAPP Aggregation Is Attenuated by the Incompletely Processed N-Terminus of proIAPP.ACS Chem Neurosci. 2022 Jul 6;13(13):2006-2016. doi: 10.1021/acschemneuro.2c00231. Epub 2022 Jun 15. ACS Chem Neurosci. 2022. PMID: 35704461 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

