Human three-dimensional in vitro model of hepatic zonation to predict zonal hepatotoxicity
- PMID: 30886645
- PMCID: PMC6404355
- DOI: 10.1186/s13036-019-0148-5
Human three-dimensional in vitro model of hepatic zonation to predict zonal hepatotoxicity
Abstract
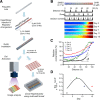
Background: Various hepatic models mimicking liver lobules have been investigated to evaluate the potential hepatotoxic effects of chemicals and drugs, but in vitro hepatic models of zonal hepatotoxicity have not yet been established. Herein, we developed a three-dimensional (3D) hepatic zonal channel to evaluate zone-specific hepatotoxicity. Based on the perivenous zone-3-like cytochrome P450 (CYP) expression patterns in metabolically active HepaRG cells treated with CHIR99021 (CHIR), which is an inducer of Wnt/β-catenin signaling, this culture model represents a novel tool for exploring hepatic zonation.
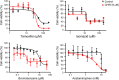
Results: We generated and validated a 3D hepatic zonal channel model in which 3D HepaRG cells were well distributed in agarose hydrogel channels, and a linear gradient of CHIR was generated according to the zonal distance. According to the results from imaging analyses and bioanalytical experiments, acetaminophen (APAP) caused cytotoxicity in the zone-3 region of the 3D hepatic zonal channel, and the levels of nonphosphorylated β-catenin, CYP2E, and apoptotic proteins were remarkably increased in the zone-3-like region. Finally, the applicability of the 3D hepatic zonal channel model for the high-throughput screening of zonal hepatotoxicity was successfully evaluated using hepatotoxic drugs, including tamoxifen, bromobenzene, and APAP.
Conclusions: The results indicated that tamoxifen induced cytotoxic effects, regardless of the zonal distance, while the zone-3-specific hepatotoxic drugs bromobenzene and APAP induced greater cytotoxic effects on cells in the zone-3-like region. This finding highlights the potential of our 3D hepatic zonation model as a valuable tool for replicating and evaluating zonal hepatotoxicity by mimicking the spatial features of liver lobules.
Keywords: Alternative hepatic model; CYP activities; Drug screening; Liver zonation; Wnt/β-catenin; Zonal hepatotoxicity.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Rappaport AM, Borowy ZJ, Lougheed WM, Lotto WN. Subdivision of hexagonal liver lobules into a structural and functional unit; role in hepatic physiology and pathology. Anat Rec. 1954;119(1):11–33. - PubMed
-
- Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. - PubMed
-
- Torre C, Perret C, Colnot S. Molecular determinants of liver zonation. Prog Mol Biol Transl Sci. 2010;97:127–150. - PubMed
-
- Jungermann K, Kietzmann T. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology. 2000;31(2):255–260. - PubMed
-
- Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol Ther. 1992;53(3):275–354. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

