Expanding the CRISPR Toolbox in Zebrafish for Studying Development and Disease
- PMID: 30886848
- PMCID: PMC6409501
- DOI: 10.3389/fcell.2019.00013
Expanding the CRISPR Toolbox in Zebrafish for Studying Development and Disease
Abstract
The study of model organisms has revolutionized our understanding of the mechanisms underlying normal development, adult homeostasis, and human disease. Much of what we know about gene function in model organisms (and its application to humans) has come from gene knockouts: the ability to show analogous phenotypes upon gene inactivation in animal models. The zebrafish (Danio rerio) has become a popular model organism for many reasons, including the fact that it is amenable to various forms of genetic manipulation. The RNA-guided CRISPR/Cas9-mediated targeted mutagenesis approaches have provided powerful tools to manipulate the genome toward developing new disease models and understanding the pathophysiology of human diseases. CRISPR-based approaches are being used for the generation of both knockout and knock-in alleles, and also for applications including transcriptional modulation, epigenome editing, live imaging of the genome, and lineage tracing. Currently, substantial effort is being made to improve the specificity of Cas9, and to expand the target coverage of the Cas9 enzymes. Novel types of naturally occurring CRISPR systems [Cas12a (Cpf1); engineered variants of Cas9, such as xCas9 and SpCas9-NG], are being studied and applied to genome editing. Since the majority of pathogenic mutations are single point mutations, development of base editors to convert C:G to T:A or A:T to G:C has further strengthened the CRISPR toolbox. In this review, we provide an overview of the increasing number of novel CRISPR-based tools and approaches, including lineage tracing and base editing.
Keywords: CRISPR/Cas9; Cas12a (Cpf1); base editors; disease models; lineage tracing; zebrafish.
Figures
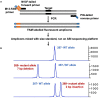

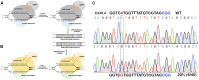
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

