An Ensemble Feature Selection Method for Biomarker Discovery
- PMID: 30887013
- PMCID: PMC6420823
- DOI: 10.1109/ISSPIT.2017.8388679
An Ensemble Feature Selection Method for Biomarker Discovery
Abstract
Feature selection in Liquid Chromatography-Mass Spectrometry (LC-MS)-based metabolomics data (biomarker discovery) have become an important topic for machine learning researchers. High dimensionality and small sample size of LC-MS data make feature selection a challenging task. The goal of biomarker discovery is to select the few most discriminative features among a large number of irreverent ones. To improve the reliability of the discovered biomarkers, we use an ensemble-based approach. Ensemble learning can improve the accuracy of feature selection by combining multiple algorithms that have complementary information. In this paper, we propose an ensemble approach to combine the results of filter-based feature selection methods. To evaluate the proposed approach, we compared it to two commonly used methods, t-test and PLS-DA, using a real data set.
Keywords: biomarker discovery; ensemble feature selection; ensemble learning; filter methods; scoring functions.
Figures

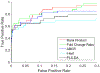

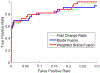
References
-
- Colburn W, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA, Woodcock J, et al., “Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. biomarkers definitions working group,” Clinical Pharmacol & Therapeutics, vol. 69, pp. 89–95, 2001. - PubMed
-
- Saeys Y, Inza I, and Larrañaga P, “A review of feature selection techniques in bioinformatics,” bioinformatics, vol. 23, no. 19, pp. 2507–2517, 2007. - PubMed
-
- Awada W, Khoshgoftaar TM, Dittman D, Wald R, and Napolitano A, “A review of the stability of feature selection techniques for bioinformatics data,” in Information Reuse and Integration (IRI), 2012 IEEE 13th International Conference on, pp. 356–363, IEEE, 2012.
-
- Lazar C, Taminau J, Meganck S, Steenhoff D, Coletta A, Molter C, de Schaetzen V, Duque R, Bersini H, and Nowe A, “A survey on filter techniques for feature selection in gene expression microarray analysis,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 9, no. 4, pp. 1106–1119, 2012. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
