Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons
- PMID: 30887021
- PMCID: PMC6487328
- DOI: 10.1093/brain/awz063
Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons
Abstract
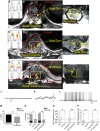
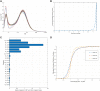
Neuropathic pain encompasses a diverse array of clinical entities affecting 7-10% of the population, which is challenging to adequately treat. Several promising therapeutics derived from molecular discoveries in animal models of neuropathic pain have failed to translate following unsuccessful clinical trials suggesting the possibility of important cellular-level and molecular differences between animals and humans. Establishing the extent of potential differences between laboratory animals and humans, through direct study of human tissues and/or cells, is likely important in facilitating translation of preclinical discoveries to meaningful treatments. Patch-clamp electrophysiology and RNA-sequencing was performed on dorsal root ganglia taken from patients with variable presence of radicular/neuropathic pain. Findings establish that spontaneous action potential generation in dorsal root ganglion neurons is associated with radicular/neuropathic pain and radiographic nerve root compression. Transcriptome analysis suggests presence of sex-specific differences and reveals gene modules and signalling pathways in immune response and neuronal plasticity related to radicular/neuropathic pain that may suggest therapeutic avenues and that has the potential to predict neuropathic pain in future cohorts.
Keywords: DRG transcriptomics; machine learning in healthcare; neuropathy; spontaneous activity.
© The Author(s) (2019). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Bannwarth B, Kostine M. Targeting nerve growth factor (NGF) for pain management: what does the future hold for NGF antagonists? Drugs 2014; 74: 619–26. - PubMed
-
- Baumann TK, Burchiel KJ, Ingram SL, Martenson ME. Responses of adult human dorsal root ganglion neurons in culture to capsaicin and low ph. Pain 1996; 65: 31–8. - PubMed
-
- Bilsky MH, Laufer I, Fourney DR, Groff M, Schmidt MH, Varga PP et al. . Reliability analysis of the epidural spinal cord compression scale. J Neurosurg Spine 2010; 13: 324–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

