Laboratory contamination over time during low-biomass sample analysis
- PMID: 30887686
- PMCID: PMC6850301
- DOI: 10.1111/1755-0998.13011
Laboratory contamination over time during low-biomass sample analysis
Abstract
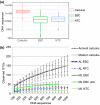
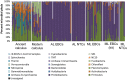
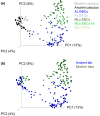
Bacteria are not only ubiquitous on earth but can also be incredibly diverse within clean laboratories and reagents. The presence of both living and dead bacteria in laboratory environments and reagents is especially problematic when examining samples with low endogenous content (e.g., skin swabs, tissue biopsies, ice, water, degraded forensic samples or ancient material), where contaminants can outnumber endogenous microorganisms within samples. The contribution of contaminants within high-throughput studies remains poorly understood because of the relatively low number of contaminant surveys. Here, we examined 144 negative control samples (extraction blank and no-template amplification controls) collected in both typical molecular laboratories and an ultraclean ancient DNA laboratory over 5 years to characterize long-term contaminant diversity. We additionally compared the contaminant content within a home-made silica-based extraction method, commonly used to analyse low endogenous content samples, with a widely used commercial DNA extraction kit. The contaminant taxonomic profile of the ultraclean ancient DNA laboratory was unique compared to modern molecular biology laboratories, and changed over time according to researcher, month and season. The commercial kit also contained higher microbial diversity and several human-associated taxa in comparison to the home-made silica extraction protocol. We recommend a minimum of two strategies to reduce the impacts of laboratory contaminants within low-biomass metagenomic studies: (a) extraction blank controls should be included and sequenced with every batch of extractions and (b) the contributions of laboratory contamination should be assessed and reported in each high-throughput metagenomic study.
Keywords: ancient DNA; contaminant; contamination; metagenomics; microbiome; microbiota.
© 2019 The Authors. Molecular Ecology Resources Published by John Wiley & Sons Ltd.
Figures

References
-
- Adler, C. J. , Dobney, K. , Weyrich, L. S. , Kaidonis, J. , Walker, A. W. , Haak, W. , … Cooper, A. (2013). Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nature Genetics, 45, 450–455. 10.1038/ng.2536 - DOI - PMC - PubMed
-
- Austin, J. J. , Smith, A. B. , Fortey, R. A. , Thomas, R. H. (1998). Ancient DNA from amber inclusions: A review of the evidence. Ancient Biomolecules, 2(2), 167–176.
-
- Barton, H. A. , Taylor, N. M. , Lubbers, B. R. , & Pemberton, A. C. (2006). DNA extraction from low‐biomass carbonate rock: An improved method with reduced contamination and the low‐biomass contaminant database. Journal of Microbiological Methods, 66, 21–31. 10.1016/j.mimet.2005.10.005 - DOI - PubMed
-
- Beckenbach, A. T. (1995). Age of bacteria from amber. Science, 270, 2015–2016; author reply 2016–2017. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

