Preservation of circadian rhythms by the protein folding chaperone, BiP
- PMID: 30888851
- PMCID: PMC6529331
- DOI: 10.1096/fj.201802366RR
Preservation of circadian rhythms by the protein folding chaperone, BiP
Abstract
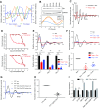
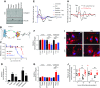
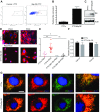
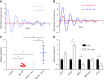
Dysregulation of collagen synthesis is associated with disease progression in cancer and fibrosis. Collagen synthesis is coordinated with the circadian clock, which in cancer cells is, curiously, deregulated by endoplasmic reticulum (ER) stress. We hypothesized interplay between circadian rhythm, collagen synthesis, and ER stress in normal cells. Here we show that fibroblasts with ER stress lack circadian rhythms in gene expression upon clock-synchronizing time cues. Overexpression of binding immunoglobulin protein (BiP) or treatment with chemical chaperones strengthens the oscillation amplitude of circadian rhythms. The significance of these findings was explored in tendon, where we showed that BiP expression is ramped preemptively prior to a surge in collagen synthesis at night, thereby preventing protein misfolding and ER stress. In turn, this forestalls activation of the unfolded protein response in order for circadian rhythms to be maintained. Thus, targeting ER stress could be used to modulate circadian rhythm and restore collagen homeostasis in disease.-Pickard, A., Chang, J., Alachkar, N., Calverley, B., Garva, R., Arvan, P., Meng, Q.-J., Kadler, K. E. Preservation of circadian rhythms by the protein folding chaperone, BiP.
Keywords: 4PBA; ER stress; Per2::luc; UDCA; collagen.
Conflict of interest statement
The authors thank P.A. and Raymond Boot-Handford (University of Michigan, Ann Arbor, MI, USA and University of Manchester, Manchester, United Kingdom, respectively) for congenital goiter–mutant thyroglobulin expression vectors. This research was funded by Wellcome Trust Investigator and Wellcome Centre Core Awards to K.E.K. (110126/Z/15/Z and 203128/Z/16/Z). P.A. was funded by U.S. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Grant DK40344. B.C. is funded by a Wellcome 4-yr Ph.D. Studentship (210062/Z/17/Z). Light microscopes in the Bioimaging Facility were additionally supported by the University of Manchester Strategic Fund. The authors declare no conflicts of interest.
Figures

References
-
- Yagita K., Tamanini F., van Der Horst G. T., Okamura H. (2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292, 278–281 - PubMed
-
- Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101, 5339–5346 - PMC - PubMed
-
- Yeung C.-Y. C., Garva R., Pickard A., Chang J., Holmes D. F., Lu Y., Mallikarjun V., Swift J., Adamson A., Calverley B., Meng Q. J., Kadler K. E. (2018) Circadian clock regulation of the secretory pathway [preprint]. bioRxiv, 304014. Retrieved May 1, 2017, from https://www.biorxiv.org/content/10.1101/304014v2 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

