Defects in assembly explain reduced antiviral activity of the G249D polymorphism in human TRIM5α
- PMID: 30889178
- PMCID: PMC6424450
- DOI: 10.1371/journal.pone.0212888
Defects in assembly explain reduced antiviral activity of the G249D polymorphism in human TRIM5α
Abstract
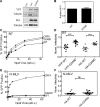
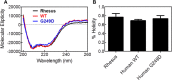
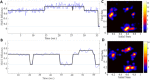
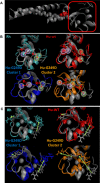
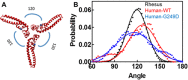
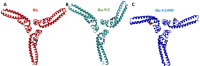
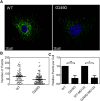
TRIM5α is an interferon inducible restriction factor which contributes to intrinsic defense against HIV infection by targeting the HIV capsid protein CA. Although human TRIM5α (huTRIM5α) does not potently inhibit HIV-1 infection, the ability of huTRIM5α to exhibit some control of HIV-1 infection is evidenced by a single nucleotide polymorphism in huTRIM5α which substitutes aspartic acid to glycine at position 249 (G249D) in the L2 region and is associated with higher susceptibility to HIV-1 infection. To understand the mechanistic basis for the reduced antiviral activity, we employed biophysical and cell biological methods coupled with molecular dynamics simulations to compare WT and the G249D polymorphism of huTRIM5α. We investigated the differences in conformational dynamics of rhesus and huTRIM5α Coiled Coil-Linker 2 (CC-L2) dimers utilizing circular dichroism and single molecule-Fluorescence Energy Transfer (sm-FRET). These methods revealed that the G249D dimer exhibits secondary structure and conformational dynamics similar to WT huTRIM5α. Homology modelling revealed that G249 was present on the hairpin of the antiparallel dimer, in a position which may act to stabilize the adjacent BBox2 domain which mediates the inter-dimeric contacts required for the formation of TRIM5 assemblies. We therefore asked if the G249D mutant forms assemblies in cells with the same efficiency as WT protein by expressing these proteins as YFP fusions and quantifying the number of assemblies in cells. In cells expressing comparable amounts of protein, the G249D mutant formed fewer assemblies than WT protein, in agreement with our homology modeling predictions and molecular dynamics simulations of dimers and higher oligomers of TRIM5α, providing a mechanistic explanation of the reduced antiviral activity of the G249D polymorphism.
Conflict of interest statement
The authors’ declare that they have no competing interests.
Figures

Similar articles
-
A naturally occurring single amino acid substitution in human TRIM5α linker region affects its anti-HIV type 1 activity and susceptibility to HIV type 1 infection.AIDS Res Hum Retroviruses. 2013 Jun;29(6):919-24. doi: 10.1089/AID.2012.0369. Epub 2013 Feb 26. AIDS Res Hum Retroviruses. 2013. PMID: 23379364 Free PMC article.
-
Restriction of HIV-1 by rhesus TRIM5α is governed by alpha helices in the Linker2 region.J Virol. 2014 Aug;88(16):8911-23. doi: 10.1128/JVI.01134-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872590 Free PMC article.
-
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article.
-
Recent insights into the mechanism and consequences of TRIM5α retroviral restriction.AIDS Res Hum Retroviruses. 2011 Mar;27(3):231-8. doi: 10.1089/AID.2010.0367. AIDS Res Hum Retroviruses. 2011. PMID: 21247355 Free PMC article. Review.
-
TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling.Curr Opin Virol. 2012 Apr;2(2):142-50. doi: 10.1016/j.coviro.2012.02.003. Epub 2012 Mar 5. Curr Opin Virol. 2012. PMID: 22482711 Free PMC article. Review.
References
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848 10.1038/nature02343 https://www.nature.com/articles/nature02343#supplementary-information. - DOI - PubMed
-
- Ozato K, Shin D-M, Chang T-H, Morse Iii HC. TRIM family proteins and their emerging roles in innate immunity. Nature Reviews Immunology. 2008;8:849 10.1038/nri2413 https://www.nature.com/articles/nri2413#supplementary-information. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

