Aicardi-Goutières Syndrome associated mutations of RNase H2B impair its interaction with ZMYM3 and the CoREST histone-modifying complex
- PMID: 30889214
- PMCID: PMC6424451
- DOI: 10.1371/journal.pone.0213553
Aicardi-Goutières Syndrome associated mutations of RNase H2B impair its interaction with ZMYM3 and the CoREST histone-modifying complex
Abstract
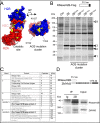
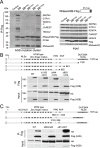
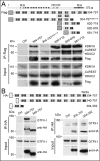
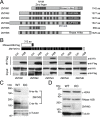
DNA-RNA hybrids arise in all cell types, and are removed by multiple enzymes, including the trimeric ribonuclease, RNase H2. Mutations in human RNase H2 result in Aicardi-Goutières syndrome (AGS), an inflammatory brain disorder notable for being a Mendelian mimic of congenital viral infection. Previous studies have shown that several AGS-associated mutations of the RNase H2B subunit do not affect trimer stability or catalytic activity and are clustered on the surface of the complex, leading us to speculate that these mutations might impair important interactions of RNase H2 with so far unidentified proteins. In this study, we show that AGS mutations in this cluster impair the interaction of RNase H2 with several members of the CoREST chromatin-silencing complex that include the histone deacetylase HDAC2 and the demethylase KDM1A, the transcriptional regulators RCOR1 and GTFII-I as well as ZMYM3, an MYM-type zinc finger protein. We also show that the interaction is mediated by the zinc finger protein ZMYM3, suggesting that ZMYM3 acts as a novel type of scaffold protein coordinating interactions between deacetylase, demethylase and RNase H type enzymes, raising the question of whether coordination between histone modifications and the degradation of RNA-DNA hybrids may be required to prevent inflammation in humans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Characterization of six recombinant human RNase H2 bearing Aicardi-Goutiéres syndrome causing mutations.J Biochem. 2019 Dec 1;166(6):537-545. doi: 10.1093/jb/mvz073. J Biochem. 2019. PMID: 31529068 Free PMC article.
-
Aicardi-Goutières syndrome: clues from the RNase H2 knock-out mouse.J Mol Med (Berl). 2013 Nov;91(11):1235-40. doi: 10.1007/s00109-013-1061-x. Epub 2013 Jun 7. J Mol Med (Berl). 2013. PMID: 23744109 Review.
-
Altered spatio-temporal dynamics of RNase H2 complex assembly at replication and repair sites in Aicardi-Goutières syndrome.Hum Mol Genet. 2014 Nov 15;23(22):5950-60. doi: 10.1093/hmg/ddu319. Epub 2014 Jun 30. Hum Mol Genet. 2014. PMID: 24986920
-
Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy.Hum Mol Genet. 2017 Oct 15;26(20):3960-3972. doi: 10.1093/hmg/ddx283. Hum Mol Genet. 2017. PMID: 29016854
-
The Balancing Act of Ribonucleotides in DNA.Trends Biochem Sci. 2016 May;41(5):434-445. doi: 10.1016/j.tibs.2016.02.005. Epub 2016 Mar 17. Trends Biochem Sci. 2016. PMID: 26996833 Free PMC article. Review.
Cited by
-
Zmym4 is required for early cranial gene expression and craniofacial cartilage formation.Front Cell Dev Biol. 2023 Oct 3;11:1274788. doi: 10.3389/fcell.2023.1274788. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37854072 Free PMC article.
-
Deleterious, protein-altering variants in the transcriptional coregulator ZMYM3 in 27 individuals with a neurodevelopmental delay phenotype.Am J Hum Genet. 2023 Feb 2;110(2):215-227. doi: 10.1016/j.ajhg.2022.12.007. Epub 2022 Dec 30. Am J Hum Genet. 2023. PMID: 36586412 Free PMC article.
-
Higher-order modular regulation of the human proteome.Mol Syst Biol. 2023 May 9;19(5):e9503. doi: 10.15252/msb.20209503. Epub 2023 Mar 9. Mol Syst Biol. 2023. PMID: 36891684 Free PMC article.
-
A CRISPR-Cas9 screen reveals genetic determinants of the cellular response to decitabine.EMBO Rep. 2025 Mar;26(6):1528-1565. doi: 10.1038/s44319-025-00385-w. Epub 2025 Feb 10. EMBO Rep. 2025. PMID: 39930152 Free PMC article.
-
Genetic and epigenetic factors shape phenotypes and outcomes in systemic lupus erythematosus - focus on juvenile-onset systemic lupus erythematosus.Curr Opin Rheumatol. 2025 Mar 1;37(2):149-163. doi: 10.1097/BOR.0000000000001072. Epub 2024 Dec 11. Curr Opin Rheumatol. 2025. PMID: 39660463 Free PMC article. Review.
References
-
- Crow YJ. Aicardi-Goutières Syndrome. Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, et al., editors. Seattle (WA): University of Washington, Seattle; 2005. - PubMed
-
- Campbell IL, Krucker T, Steffensen S, Akwa Y, Powell HC, Lane T, et al. Structural and functional neuropathology in transgenic mice with CNS expression of IFN-alpha. Brain Res. 1999;835: 46–61. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

