Machine learning in medicine: a practical introduction
- PMID: 30890124
- PMCID: PMC6425557
- DOI: 10.1186/s12874-019-0681-4
Machine learning in medicine: a practical introduction
Abstract
Background: Following visible successes on a wide range of predictive tasks, machine learning techniques are attracting substantial interest from medical researchers and clinicians. We address the need for capacity development in this area by providing a conceptual introduction to machine learning alongside a practical guide to developing and evaluating predictive algorithms using freely-available open source software and public domain data.
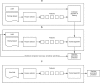
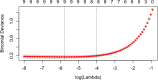
Methods: We demonstrate the use of machine learning techniques by developing three predictive models for cancer diagnosis using descriptions of nuclei sampled from breast masses. These algorithms include regularized General Linear Model regression (GLMs), Support Vector Machines (SVMs) with a radial basis function kernel, and single-layer Artificial Neural Networks. The publicly-available dataset describing the breast mass samples (N=683) was randomly split into evaluation (n=456) and validation (n=227) samples. We trained algorithms on data from the evaluation sample before they were used to predict the diagnostic outcome in the validation dataset. We compared the predictions made on the validation datasets with the real-world diagnostic decisions to calculate the accuracy, sensitivity, and specificity of the three models. We explored the use of averaging and voting ensembles to improve predictive performance. We provide a step-by-step guide to developing algorithms using the open-source R statistical programming environment.
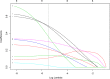
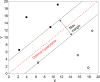
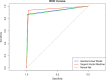
Results: The trained algorithms were able to classify cell nuclei with high accuracy (.94 -.96), sensitivity (.97 -.99), and specificity (.85 -.94). Maximum accuracy (.96) and area under the curve (.97) was achieved using the SVM algorithm. Prediction performance increased marginally (accuracy =.97, sensitivity =.99, specificity =.95) when algorithms were arranged into a voting ensemble.
Conclusions: We use a straightforward example to demonstrate the theory and practice of machine learning for clinicians and medical researchers. The principals which we demonstrate here can be readily applied to other complex tasks including natural language processing and image recognition.
Keywords: Classification; Computer-assisted; Decision making; Diagnosis; Medical informatics; Programming languages; Supervised machine learning.
Conflict of interest statement
Ethics approval and consent to participate
In this manuscript we use de-identified data from a public repository [17]. The data are included on the
Consent for publication
All contributing parties consent for the publication of this work.
Competing interests
The authors report no competing interests relating to this work.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

















References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

