The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila
- PMID: 30890568
- PMCID: PMC6467472
- DOI: 10.1242/dev.175570
The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila
Abstract
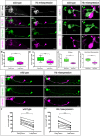
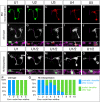
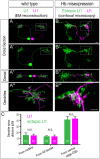
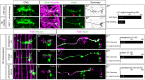
The generation of neuronal diversity is essential for circuit formation and behavior. Morphological differences in sequentially born neurons could be due to intrinsic molecular identity specified by temporal transcription factors (henceforth called intrinsic temporal identity) or due to changing extrinsic cues. Here, we have used the Drosophila NB7-1 lineage to address this issue. NB7-1 generates the U1-U5 motor neurons sequentially; each has a distinct intrinsic temporal identity due to inheritance of different temporal transcription factors at its time of birth. We show that the U1-U5 neurons project axons sequentially, followed by sequential dendrite extension. We misexpressed the earliest temporal transcription factor, Hunchback, to create 'ectopic' U1 neurons with an early intrinsic temporal identity but later birth-order. These ectopic U1 neurons have axon muscle targeting and dendrite neuropil targeting that are consistent with U1 intrinsic temporal identity, rather than with their time of birth or differentiation. We conclude that intrinsic temporal identity plays a major role in establishing both motor axon muscle targeting and dendritic arbor targeting, which are required for proper motor circuit development.
Keywords: Dendrite morphology; Heterochronic; Hunchback; Larval locomotion; Motor circuits; Motor neuron; Neural circuits; Temporal identity; Temporal transcription factors.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
The Hunchback transcription factor determines interneuron molecular identity, morphology, and presynapse targeting in the Drosophila NB5-2 lineage.PLoS Biol. 2025 Mar 31;23(3):e3002881. doi: 10.1371/journal.pbio.3002881. eCollection 2025 Mar. PLoS Biol. 2025. PMID: 40163536 Free PMC article.
-
Imp and Chinmo are required for embryonic motor neuron axon and dendrite targeting.Biol Open. 2025 Jul 15;14(7):bio062105. doi: 10.1242/bio.062105. Epub 2025 Jul 25. Biol Open. 2025. PMID: 40720095 Free PMC article.
-
The Hunchback temporal transcription factor establishes, but is not required to maintain, early-born neuronal identity.Neural Dev. 2017 Jan 31;12(1):1. doi: 10.1186/s13064-017-0078-1. Neural Dev. 2017. PMID: 28137283 Free PMC article.
-
From temporal patterning to neuronal connectivity in Drosophila type I neuroblast lineages.Semin Cell Dev Biol. 2023 Jun;142:4-12. doi: 10.1016/j.semcdb.2022.05.022. Epub 2022 May 31. Semin Cell Dev Biol. 2023. PMID: 35659165 Free PMC article. Review.
-
Cell-intrinsic drivers of dendrite morphogenesis.Development. 2013 Dec;140(23):4657-71. doi: 10.1242/dev.087676. Development. 2013. PMID: 24255095 Free PMC article. Review.
Cited by
-
Drosophila Embryonic CNS Development: Neurogenesis, Gliogenesis, Cell Fate, and Differentiation.Genetics. 2019 Dec;213(4):1111-1144. doi: 10.1534/genetics.119.300974. Genetics. 2019. PMID: 31796551 Free PMC article. Review.
-
How prolonged expression of Hunchback, a temporal transcription factor, re-wires locomotor circuits.Elife. 2019 Sep 10;8:e46089. doi: 10.7554/eLife.46089. Elife. 2019. PMID: 31502540 Free PMC article.
-
Integrated Patterning Programs During Drosophila Development Generate the Diversity of Neurons and Control Their Mature Properties.Annu Rev Neurosci. 2021 Jul 8;44:153-172. doi: 10.1146/annurev-neuro-102120-014813. Epub 2021 Feb 8. Annu Rev Neurosci. 2021. PMID: 33556251 Free PMC article.
-
The VAPB Axis Precisely Coordinates the Timing of Motoneuron Dendritogenesis in Neural Map Development.Res Sq [Preprint]. 2024 Dec 31:rs.3.rs-5684747. doi: 10.21203/rs.3.rs-5684747/v1. Res Sq. 2024. PMID: 39801516 Free PMC article. Preprint.
-
Presynaptic developmental plasticity allows robust sparse wiring of the Drosophila mushroom body.Elife. 2020 Jan 8;9:e52278. doi: 10.7554/eLife.52278. Elife. 2020. PMID: 31913123 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

