More Evidence of Collusion: a New Prophage-Mediated Viral Defense System Encoded by Mycobacteriophage Sbash
- PMID: 30890613
- PMCID: PMC6426596
- DOI: 10.1128/mBio.00196-19
More Evidence of Collusion: a New Prophage-Mediated Viral Defense System Encoded by Mycobacteriophage Sbash
Abstract
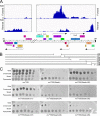
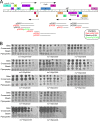
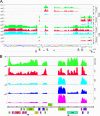
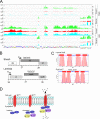
The arms race between bacteria and their bacteriophages profoundly influences microbial evolution. With an estimated 1023 phage infections occurring per second, there is strong selection for both bacterial survival and phage coevolution for continued propagation. Many phage resistance systems, including restriction-modification systems, clustered regularly interspaced short palindromic repeat-Cas (CRISPR-Cas) systems, a variety of abortive infection systems, and many others that are not yet mechanistically defined, have been described. Temperate bacteriophages are common and form stable lysogens that are immune to superinfection by the same or closely related phages. However, temperate phages collude with their hosts to confer defense against genomically distinct phages, to the mutual benefit of the bacterial host and the prophage. Prophage-mediated viral systems have been described in Mycobacterium phages and Pseudomonas phages but are predicted to be widespread throughout the microbial world. Here we describe a new viral defense system in which the mycobacteriophage Sbash prophage colludes with its Mycobacterium smegmatis host to confer highly specific defense against infection by the unrelated mycobacteriophage Crossroads. Sbash genes 30 and 31 are lysogenically expressed and are necessary and sufficient to confer defense against Crossroads but do not defend against any of the closely related phages grouped in subcluster L2. The mapping of Crossroads defense escape mutants shows that genes 132 and 141 are involved in recognition by the Sbash defense system and are proposed to activate a loss in membrane potential mediated by Sbash gp30 and gp31.IMPORTANCE Viral infection is an ongoing challenge to bacterial survival, and there is strong selection for development or acquisition of defense systems that promote survival when bacteria are attacked by bacteriophages. Temperate phages play central roles in these dynamics through lysogenic expression of genes that defend against phage attack, including those unrelated to the prophage. Few prophage-mediated viral defense systems have been characterized, but they are likely widespread both in phage genomes and in the prophages integrated in bacterial chromosomes.
Keywords: Mycobacterium; bacteriophage; viral defense.
Copyright © 2019 Gentile et al.
Figures





References
-
- Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, Modlin RL, Hendrix RW, Hatfull GF. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi:10.1016/j.virol.2012.09.026. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
