Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis
- PMID: 30890697
- PMCID: PMC6424977
- DOI: 10.1038/s41467-019-09182-1
Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis
Abstract
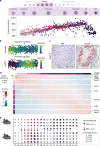
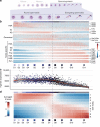
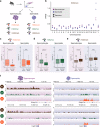
Male gametes are generated through a specialised differentiation pathway involving a series of developmental transitions that are poorly characterised at the molecular level. Here, we use droplet-based single-cell RNA-Sequencing to profile spermatogenesis in adult animals and at multiple stages during juvenile development. By exploiting the first wave of spermatogenesis, we both precisely stage germ cell development and enrich for rare somatic cell-types and spermatogonia. To capture the full complexity of spermatogenesis including cells that have low transcriptional activity, we apply a statistical tool that identifies previously uncharacterised populations of leptotene and zygotene spermatocytes. Focusing on post-meiotic events, we characterise the temporal dynamics of X chromosome re-activation and profile the associated chromatin state using CUT&RUN. This identifies a set of genes strongly repressed by H3K9me3 in spermatocytes, which then undergo extensive chromatin remodelling post-meiosis, thus acquiring an active chromatin state and spermatid-specific expression.
Conflict of interest statement
The Authors declare no Competing Interests.
Figures





Similar articles
-
Distribution of the sex chromosome during mouse spermatogenesis in testis tissue sections.J Reprod Dev. 2015;61(5):375-81. doi: 10.1262/jrd.2015-013. Epub 2015 Jun 14. J Reprod Dev. 2015. PMID: 26073979 Free PMC article.
-
The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids.BMC Genomics. 2010 Jun 10;11:367. doi: 10.1186/1471-2164-11-367. BMC Genomics. 2010. PMID: 20537150 Free PMC article.
-
Expression and epigenomic landscape of the sex chromosomes in mouse post-meiotic male germ cells.Epigenetics Chromatin. 2016 Oct 27;9:47. doi: 10.1186/s13072-016-0099-8. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27795737 Free PMC article.
-
Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes.BMC Genomics. 2016 Aug 12;17(1):628. doi: 10.1186/s12864-016-2865-1. BMC Genomics. 2016. PMID: 27519264 Free PMC article.
-
Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression.Bioessays. 2012 Mar;34(3):187-93. doi: 10.1002/bies.201100141. Epub 2011 Dec 15. Bioessays. 2012. PMID: 22170506 Review.
Cited by
-
Efficient low-cost chromatin profiling with CUT&Tag.Nat Protoc. 2020 Oct;15(10):3264-3283. doi: 10.1038/s41596-020-0373-x. Epub 2020 Sep 10. Nat Protoc. 2020. PMID: 32913232 Free PMC article.
-
Characterization of the protein expression and localization of hnRNP family members during murine spermatogenesis.Asian J Androl. 2023 May-Jun;25(3):314-321. doi: 10.4103/aja202273. Asian J Androl. 2023. PMID: 36124536 Free PMC article.
-
Single-Cell RNA Sequencing Reveals an Atlas of Hezuo Pig Testis Cells.Int J Mol Sci. 2024 Sep 10;25(18):9786. doi: 10.3390/ijms25189786. Int J Mol Sci. 2024. PMID: 39337274 Free PMC article.
-
Meiosis-specific ZFP541 repressor complex promotes developmental progression of meiotic prophase towards completion during mouse spermatogenesis.Nat Commun. 2021 Jun 1;12(1):3184. doi: 10.1038/s41467-021-23378-4. Nat Commun. 2021. PMID: 34075040 Free PMC article.
-
Single-cell RNA-sequencing reveals the dynamic process and novel markers in porcine spermatogenesis.J Anim Sci Biotechnol. 2021 Dec 7;12(1):122. doi: 10.1186/s40104-021-00638-3. J Anim Sci Biotechnol. 2021. PMID: 34872612 Free PMC article.
References
-
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000;21:776–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

