Identification of Gene Expression Changes Associated With Uterine Receptivity in Mice
- PMID: 30890945
- PMCID: PMC6413723
- DOI: 10.3389/fphys.2019.00125
Identification of Gene Expression Changes Associated With Uterine Receptivity in Mice
Abstract
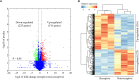
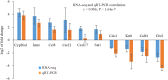
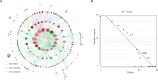
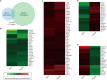
The mouse is a widely used animal model for studying human reproduction. Although global gene expression changes associated with human uterine receptivity have been determined by independent groups, the same studies in the mouse are scarce. The extent of similarities/differences between mice and humans on uterine receptivity at the molecular level remains to be determined. In the present study, we analyzed global gene expression changes in receptive uterus on day 4 of pregnancy compared to non-receptive uterus on day 3 of pregnancy in mice. A total of 541 differentially expressed genes were identified, of which 316 genes were up-regulated and 225 genes were down-regulated in receptive uterus compared to non-receptive uterus. Gene ontology and gene network analysis highlighted the activation of inflammatory response in the receptive uterus. By analyzing the promoter sequences of differentially expressed genes, we identified 12 causal transcription factors. Through connectivity map (CMap) analysis, we revealed several compounds with potential anti-receptivity activity. Finally, we performed a cross-species comparison against human uterine receptivity from a published dataset. Our study provides a valuable resource for understanding the molecular mechanism underlying uterine receptivity in mice.
Keywords: RNA-seq; gene expression; mouse; receptivity; uterus.
Figures



References
-
- Altmae S., Reimand J., Hovatta O., Zhang P., Kere J., Laisk T., et al. (2012). Research resource: interactome of human embryo implantation: identification of gene expression pathways, regulation, and integrated regulatory networks. Mol. Endocrinol. 26 203–217. 10.1210/me.2011-1196 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

