Primary Vitamin D Target Genes of Human Monocytes
- PMID: 30890957
- PMCID: PMC6411690
- DOI: 10.3389/fphys.2019.00194
Primary Vitamin D Target Genes of Human Monocytes
Abstract
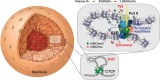
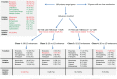
The molecular basis of vitamin D signaling implies that the metabolite 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) of the secosteroid vitamin D3 activates the transcription factor vitamin D receptor (VDR), which in turn modulates the expression of hundreds of primary vitamin D target genes. Since the evolutionary role of nuclear receptors, such as VDR, was the regulation of cellular metabolism, the control of calcium metabolism became the primary function of vitamin D and its receptor. Moreover, the nearly ubiquitous expression of VDR enabled vitamin D to acquire additional physiological functions, such as the support of the innate immune system in its defense against microbes. Monocytes and their differentiated phenotypes, macrophages and dendritic cells, are key cell types of the innate immune system. Vitamin D signaling was most comprehensively investigated in THP-1 cells, which are an established model of human monocytes. This includes the 1,25(OH)2D3-modulated cistromes of VDR, the pioneer transcription factors PU.1 and CEBPA and the chromatin modifier CTCF as well as of the histone markers of promoter and enhancer regions, H3K4me3 and H3K27ac, respectively. These epigenome-wide datasets led to the development of our chromatin model of vitamin D signaling. This review discusses the mechanistic basis of 189 primary vitamin D target genes identified by transcriptome-wide analysis of 1,25(OH)2D3-stimulated THP-1 cells and relates the epigenomic basis of four different regulatory scenarios to the physiological functions of the respective genes.
Keywords: VDR; epigenome; gene regulation; monocytes; transcriptome; vitamin D; vitamin D target genes.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources

