Structural Insights Into Key Plasmodium Proteases as Therapeutic Drug Targets
- PMID: 30891019
- PMCID: PMC6411711
- DOI: 10.3389/fmicb.2019.00394
Structural Insights Into Key Plasmodium Proteases as Therapeutic Drug Targets
Abstract
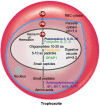
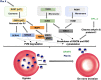
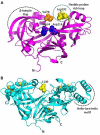
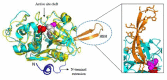
Malaria, caused by protozoan of genus Plasmodium, remains one of the highest mortality infectious diseases. Malaria parasites have a complex life cycle, easily adapt to their host's immune system and have evolved with an arsenal of unique proteases which play crucial roles in proliferation and survival within the host cells. Owing to the existing knowledge of enzymatic mechanisms, 3D structures and active sites of proteases, they have been proven to be opportune for target based drug development. Here, we discuss in depth the crucial roles of essential proteases in Plasmodium life cycle and particularly focus on highlighting the atypical "structural signatures" of key parasite proteases which have been exploited for drug development. These features, on one hand aid parasites pathogenicity while on the other hand could be effective in designing targeted and very specific inhibitors for counteracting them. We conclude that Plasmodium proteases are suitable as multistage targets for designing novel drugs with new modes of action to combat malaria.
Keywords: Ca2+-dependent subtilase; aspartyl protease fold; drug targets; malaria; papain-family cysteine proteases; proteases; therapeutics.
Figures
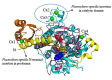

References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

