Application of Duplex Fluorescence Melting Curve Analysis (FMCA) to Identify Canine Parvovirus Type 2 Variants
- PMID: 30891024
- PMCID: PMC6411689
- DOI: 10.3389/fmicb.2019.00419
Application of Duplex Fluorescence Melting Curve Analysis (FMCA) to Identify Canine Parvovirus Type 2 Variants
Abstract
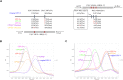
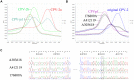
Canine parvovirus (CPV-2) is an enteric virus causing morbidity and mortality in dogs worldwide. Since CPV-2 emerged as canine pathogen, the original CPV-2 strain has constantly evolved, and its primary variants (CPV-2a, CPV-2b, and CPV-2c) co-circulate to varying extents in canine populations worldwide. Thus, rapid and accurate laboratory diagnoses of CPV-2 variants are crucial to monitor CPV-2 evolution. Conventional methods for CPV-2 genotyping are laborious, time consuming, and determining the genotype of a CPV-2 variant often requires two or more reaction tubes. The present study developed a probe-based fluorescence melting curve analysis (FMCA) for genotyping six different CPV-2 variants (original CPV-2, CPV-2a, CPV-2b, CPV-2c, and vaccine strains of CPVpf and CPVint) in a single reaction tube using only two TaqMan probes. One of the TaqMan probes (FAM labeled) was designed to perfectly match with the target sequence of CPV-2a, this probe allows a 1-bp mismatched hybridization with the CPV-2b VP2 gene region (A4062G), and a 2-bp mismatched hybridization for CPV-2c (A4062G and T4064A); Another TaqMan probe (HEX labeled) was produced to perfectly match with the target sequence of original CPV-2, this probe enables 1-bp mismatched hybridization with the other CPV-2 variants (A3045T). Using the two TaqMan probes, all six CPV-2 variants were readily distinguished by their respective melting temperature values in a single reaction tube. The detection limits of this assay were 1-10 copies per reaction for six CPV-2 construction plasmids and no cross reactions were observed with several other common canine viruses. In this assay, co-infected samples were also directly identified via probe-based FMCA without using a mixing control; only a pure control is required. The clinical evaluation of this assay was demonstrated by analyzing 83 clinical fecal samples, among which 41 (49.39%), 8 (9.63%), and 14 (16.87%) samples were found to be positive for CPV-2a, CPV-2b, and CPV-2c, respectively. The concordance rate between probe-based FMCA and Sanger sequencing was 100%. Thus, the duplex FMCA is effective, rapid, simple, high-throughput, and straightforward for genotyping CPV-2 variants, and is useful to effectively diagnose and monitor CPV-2 epidemiology.
Keywords: CPV-2 variant; TaqMan probe; genotyping; melting curve; sequencing.
Figures
References
LinkOut - more resources
Full Text Sources

