The Formation of Glycan-Specific Natural Antibodies Repertoire in GalT-KO Mice Is Determined by Gut Microbiota
- PMID: 30891034
- PMCID: PMC6411795
- DOI: 10.3389/fimmu.2019.00342
The Formation of Glycan-Specific Natural Antibodies Repertoire in GalT-KO Mice Is Determined by Gut Microbiota
Abstract
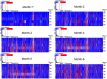
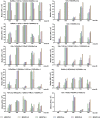
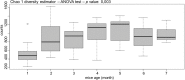
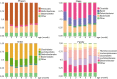
Gut commensal bacteria are known to have a significant role in regulating the innate and adaptive immune homeostasis. Alterations in the intestinal microbial composition have been associated with several disease states, including autoimmune and inflammatory conditions. However, it is not entirely clear how commensal gut microbiota modulate and contribute to the systemic immunity, and whether circulating elements of the host immune system could regulate the microbiome. Thus, we have studied the diversity and abundance of specific taxons in the gut microbiota of inbred GalT-KO mice during 7 months of animal life by metagenetic high-throughput sequencing (16S rRNA gene, variable regions V3-V5). The repertoire of glycan-specific natural antibodies, obtained by printed glycan array technology, was then associated with the microbial diversity for each animal by metagenome-wide association studies (MWAS). Our data show that the orders clostridiales (most abundant), bacteriodales, lactobacillales, and deferribacterales may be associated with the development of the final repertoire of natural anti-glycan antibodies in GalT-KO mice. The main changes in microbiota diversity (month-2 and month-3) were related to important changes in levels and repertoire of natural anti-glycan antibodies in these mice. Additionally, significant positive and negative associations were found between the gut microbiota and the pattern of specific anti-glycan antibodies. Regarding individual features, the gut microbiota and the corresponding repertoire of natural anti-glycan antibodies showed differences among the examined animals. We also found redundancy in different taxa associated with the development of specific anti-glycan antibodies. Differences in microbial diversity did not, therefore, necessarily influence the overall functional output of the gut microbiome of GalT-KO mice. In summary, the repertoire of natural anti-carbohydrate antibodies may be partially determined by the continuous antigenic stimulation produced by the gut bacterial population of each GalT-KO mouse. Small differences in gut microbiota diversity could determine different repertoire and levels of natural anti-glycan antibodies and consequently might induce different immune responses to pathogens or other potential threats.
Keywords: 16S rRNA gene; GalT-KO mice; gut microbiota; metagenetic high-throughput sequencing; metagenome-wide association studies; natural anti-glycan antibodies; printed glycan array.
Figures

Similar articles
-
The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection.Microbiol Spectr. 2015 Jun;3(3). doi: 10.1128/microbiolspec.MBP-0008-2014. Microbiol Spectr. 2015. PMID: 26185088
-
Impact of a 3-Months Vegetarian Diet on the Gut Microbiota and Immune Repertoire.Front Immunol. 2018 Apr 27;9:908. doi: 10.3389/fimmu.2018.00908. eCollection 2018. Front Immunol. 2018. PMID: 29755475 Free PMC article.
-
Specificity profile of αGal antibodies in αGalT KO mice as probed with comprehensive printed glycan array: Comparison with human anti-Galili antibodies.Xenotransplantation. 2021 May;28(3):e12672. doi: 10.1111/xen.12672. Epub 2021 Jan 12. Xenotransplantation. 2021. PMID: 33432698
-
Manipulation of the glycan-specific natural antibody repertoire for immunotherapy.Immunol Rev. 2016 Mar;270(1):32-50. doi: 10.1111/imr.12397. Immunol Rev. 2016. PMID: 26864103 Free PMC article. Review.
-
Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex.J Autoimmun. 2018 Aug;92:12-34. doi: 10.1016/j.jaut.2018.05.008. Epub 2018 Jun 1. J Autoimmun. 2018. PMID: 29861127 Review.
Cited by
-
Long-distance relationships - regulation of systemic host defense against infections by the gut microbiota.Mucosal Immunol. 2022 May;15(5):809-818. doi: 10.1038/s41385-022-00539-2. Epub 2022 Jun 22. Mucosal Immunol. 2022. PMID: 35732817 Review.
-
Emergence and significance of carbohydrate-specific antibodies.Genes Immun. 2020 Aug;21(4):224-239. doi: 10.1038/s41435-020-0105-9. Epub 2020 Aug 5. Genes Immun. 2020. PMID: 32753697 Free PMC article. Review.
-
Development of smart anti-glycan reagents using immunized lampreys.Commun Biol. 2020 Feb 28;3(1):91. doi: 10.1038/s42003-020-0819-2. Commun Biol. 2020. PMID: 32111965 Free PMC article.
-
Mechanisms of Formation of Antibodies against Blood Group Antigens That Do Not Exist in the Body.Int J Mol Sci. 2023 Oct 10;24(20):15044. doi: 10.3390/ijms242015044. Int J Mol Sci. 2023. PMID: 37894724 Free PMC article.
-
ABO Blood Types and COVID-19: Spurious, Anecdotal, or Truly Important Relationships? A Reasoned Review of Available Data.Viruses. 2021 Jan 22;13(2):160. doi: 10.3390/v13020160. Viruses. 2021. PMID: 33499228 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

