Current Advances on Structure-Function Relationships of Pyridoxal 5'-Phosphate-Dependent Enzymes
- PMID: 30891451
- PMCID: PMC6411801
- DOI: 10.3389/fmolb.2019.00004
Current Advances on Structure-Function Relationships of Pyridoxal 5'-Phosphate-Dependent Enzymes
Abstract
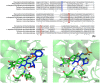
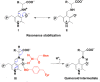
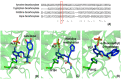
Pyridoxal 5'-phosphate (PLP) functions as a coenzyme in many enzymatic processes, including decarboxylation, deamination, transamination, racemization, and others. Enzymes, requiring PLP, are commonly termed PLP-dependent enzymes, and they are widely involved in crucial cellular metabolic pathways in most of (if not all) living organisms. The chemical mechanisms for PLP-mediated reactions have been well elaborated and accepted with an emphasis on the pure chemical steps, but how the chemical steps are processed by enzymes, especially by functions of active site residues, are not fully elucidated. Furthermore, the specific mechanism of an enzyme in relation to the one for a similar class of enzymes seems scarcely described or discussed. This discussion aims to link the specific mechanism described for the individual enzyme to the same types of enzymes from different species with aminotransferases, decarboxylases, racemase, aldolase, cystathionine β-synthase, aromatic phenylacetaldehyde synthase, et al. as models. The structural factors that contribute to the reaction mechanisms, particularly active site residues critical for dictating the reaction specificity, are summarized in this review.
Keywords: amino acid residues; pyridoxal 5′-phosphate; reaction mechanism; reaction specificity; structure-function relationship.
Figures





References
-
- Beattie A. E., Clarke D. J., Wadsworth J. M., Lowther J., Sin H. L., Campopiano D. J. (2013). Reconstitution of the pyridoxal 5′-phosphate (PLP) dependent enzyme serine palmitoyltransferase (SPT) with pyridoxal reveals a crucial role for the phosphate during catalysis. Chem. Commun. 49, 7058–7060. 10.1039/c3cc43001d - DOI - PubMed
-
- Berkovitch F., Behshad E., Tang K. H., Enns E. A., Frey P. A., Drennan C. L. (2004). A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase. Proc. Natl. Acad. Sci. U.S.A. 101, 15870–15875. 10.1073/pnas.0407074101 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

