Cardiac glycosides decrease influenza virus replication by inhibiting cell protein translational machinery
- PMID: 30892074
- PMCID: PMC6620673
- DOI: 10.1152/ajplung.00173.2018
Cardiac glycosides decrease influenza virus replication by inhibiting cell protein translational machinery
Abstract
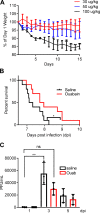
Cardiac glycosides (CGs) are used primarily for cardiac failure and have been reported to have other effects, including inhibition of viral replication. Here we set out to study mechanisms by which CGs as inhibitors of the Na-K-ATPase decrease influenza A virus (IAV) replication in the lungs. We found that CGs inhibit influenza virus replication in alveolar epithelial cells by decreasing intracellular potassium, which in turn inhibits protein translation, independently of viral entry, mRNA transcription, and protein degradation. These effects were independent of the Src signaling pathway and intracellular calcium concentration changes. We found that short-term treatment with ouabain prevented IAV replication without cytotoxicity. Rodents express a Na-K-ATPase-α1 resistant to CGs. Thus we utilized Na-K-ATPase-α1-sensitive mice, infected them with high doses of influenza virus, and observed a modest survival benefit when treated with ouabain. In summary, we provide evidence that the inhibition of the Na-K-ATPase by CGs decreases influenza A viral replication by modulating the cell protein translational machinery and results in a modest survival benefit in mice.
Keywords: Na-K-ATPase; antiviral treatment; intracellular potassium.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures
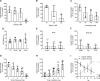

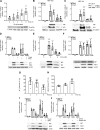
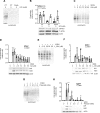
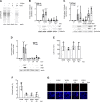
References
-
- Bertorello AM, Komarova Y, Smith K, Leibiger IB, Efendiev R, Pedemonte CH, Borisy G, Sznajder JI. Analysis of Na+,K+-ATPase motion and incorporation into the plasma membrane in response to G protein-coupled receptor signals in living cells. Mol Biol Cell 14: 1149–1157, 2003. doi:10.1091/mbc.e02-06-0367. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

